Please be patient as the page loads
|
PKHF1_MOUSE
|
||||||
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PKHF1_MOUSE
|
||||||
| θ value | 1.86219e-138 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
PKHF1_HUMAN
|
||||||
| θ value | 3.4095e-124 (rank : 2) | NC score | 0.998644 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
PKHF2_MOUSE
|
||||||
| θ value | 2.24094e-75 (rank : 3) | NC score | 0.980578 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91WB4 | Gene names | Plekhf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2). | |||||
|
PKHF2_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 4) | NC score | 0.980969 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H8W4 | Gene names | PLEKHF2, ZFYVE18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2) (PH and FYVE domain-containing protein 2) (Phafin-2) (Zinc finger FYVE domain-containing protein 18). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 5) | NC score | 0.731069 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 6) | NC score | 0.715782 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 7) | NC score | 0.716356 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 8) | NC score | 0.724888 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
FGD2_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 9) | NC score | 0.643042 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
FGD4_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 10) | NC score | 0.619085 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD2_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 11) | NC score | 0.638950 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 12) | NC score | 0.613406 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 13) | NC score | 0.604669 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 14) | NC score | 0.601076 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 15) | NC score | 0.723896 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 16) | NC score | 0.246798 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 17) | NC score | 0.228319 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
WDFY3_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 18) | NC score | 0.408492 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
WDFY3_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 19) | NC score | 0.409661 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 20) | NC score | 0.670830 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
ZFY16_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 21) | NC score | 0.642571 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
ZFY16_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 22) | NC score | 0.652996 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 23) | NC score | 0.398531 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 24) | NC score | 0.316162 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 25) | NC score | 0.391388 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 26) | NC score | 0.317447 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 27) | NC score | 0.533681 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 28) | NC score | 0.396840 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 29) | NC score | 0.531548 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
ZFY21_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 30) | NC score | 0.702290 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 31) | NC score | 0.315443 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
ZFY21_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 32) | NC score | 0.705676 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 33) | NC score | 0.269172 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 34) | NC score | 0.389431 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 35) | NC score | 0.268772 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 36) | NC score | 0.556525 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 37) | NC score | 0.309017 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 38) | NC score | 0.470146 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 39) | NC score | 0.469369 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
ZFYV1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 40) | NC score | 0.559712 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 41) | NC score | 0.257899 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
WDFY1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 42) | NC score | 0.417867 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IWB7, Q9H9D5, Q9P2B3 | Gene names | WDFY1, KIAA1435, WDF1, ZFYVE17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 1 (WD40- and FYVE domain- containing protein 1) (Phosphoinositide-binding protein 1) (FENS-1) (Zinc finger FYVE domain-containing protein 17). | |||||
|
ZFYV1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 43) | NC score | 0.554732 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
WDFY2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 44) | NC score | 0.392718 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96P53, Q96CS1 | Gene names | WDFY2, WDF2, ZFYVE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2) (Zinc finger FYVE domain-containing protein 22). | |||||
|
WDFY2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 45) | NC score | 0.396321 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BUB4 | Gene names | Wdfy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 46) | NC score | 0.198089 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 47) | NC score | 0.486577 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
RBNS5_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 48) | NC score | 0.480288 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
FGD3_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 49) | NC score | 0.514413 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 50) | NC score | 0.186109 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 51) | NC score | 0.229848 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
ZFY19_HUMAN
|
||||||
| θ value | 0.279714 (rank : 52) | NC score | 0.408506 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
ZFY19_MOUSE
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.402186 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DAZ9, Q8VCV7 | Gene names | Zfyve19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19. | |||||
|
PREX1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.251027 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.093608 (rank : 104) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.088668 (rank : 108) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.039897 (rank : 141) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.040609 (rank : 139) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
MELPH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.063784 (rank : 126) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.016551 (rank : 151) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.065003 (rank : 124) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.035011 (rank : 144) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
EXOC8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.040419 (rank : 140) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGF7 | Gene names | Exoc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 8 (Exocyst complex 84 kDa subunit). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.035850 (rank : 143) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
S4A7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.034365 (rank : 145) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6M7, O60350, Q6AHZ9, Q9HC88, Q9UIB9 | Gene names | SLC4A7, BT, NBC2, NBC2B, NBC3, SBC2, SLC4A6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium bicarbonate cotransporter 3 (Sodium bicarbonate cotransporter 2) (Sodium bicarbonate cotransporter 2b) (Bicarbonate transporter) (Solute carrier family 4 member 7). | |||||
|
S4A7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.035880 (rank : 142) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BTY2, Q3U5H7 | Gene names | Slc4a7, Nbc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium bicarbonate cotransporter 3 (Solute carrier family 4 member 7). | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.015039 (rank : 152) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.058109 (rank : 132) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
RFFL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.103482 (rank : 99) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
S4A10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.028927 (rank : 149) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6U841, Q4ZFX6, Q8TCP2, Q9HCQ6 | Gene names | SLC4A10, NCBE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-driven chloride bicarbonate exchanger (Solute carrier family 4 member 10). | |||||
|
S4A10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.029103 (rank : 147) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DTL9, Q8CFS3, Q9EST0 | Gene names | Slc4a10, Kiaa4136, Ncbe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-driven chloride bicarbonate exchanger (Solute carrier family 4 member 10). | |||||
|
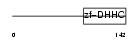
ZDH21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.020118 (rank : 150) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D270, Q6EMK1, Q80XQ3 | Gene names | Zdhhc21, Gramp3 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21) (GABA-A receptor-associated membrane protein 3). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.014206 (rank : 153) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.006196 (rank : 155) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.059437 (rank : 131) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
RNF34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.103390 (rank : 100) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.030733 (rank : 146) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.029061 (rank : 148) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.013542 (rank : 154) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
TRI14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.003353 (rank : 156) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVW3, Q6A0C3, Q762I6, Q9D3G8 | Gene names | Trim14, Kiaa0129, Pub | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 14 (PU.1-binding protein). | |||||
|
ABR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.140846 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
ARHG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.118855 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.117378 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
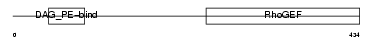
ARHG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.107042 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92974, O75142, Q15079 | Gene names | ARHGEF2, KIAA0651, LFP40 | |||
|
Domain Architecture |
|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (GEF-H1 protein) (Proliferating cell nucleolar antigen p40). | |||||
|
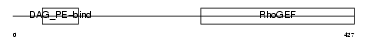
ARHG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.156732 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60875, O09115 | Gene names | Arhgef2, Lbcl1, Lfc | |||
|
Domain Architecture |
|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (Lymphoid blast crisis- like 1) (LBC'S first cousin) (Oncogene LFC) (RHOBIN). | |||||
|
ARHG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.080706 (rank : 117) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
ARHG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.081990 (rank : 114) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91X46, Q8CDM0, Q91VY4, Q99K14, Q9DC31 | Gene names | Arhgef3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3. | |||||
|
ARHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.155659 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.155247 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.180570 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
ARHG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.076356 (rank : 120) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
ARHG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.069305 (rank : 123) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.118857 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
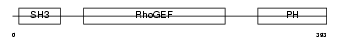
ARHG7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.097118 (rank : 103) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |
|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
ARHG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.101748 (rank : 102) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z628, Q12773, Q96D82, Q99903, Q9UEN6 | Gene names | NET1, ARHGEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroepithelial cell-transforming gene 1 protein (p65 Net1 proto- oncogene) (Rho guanine nucleotide exchange factor 8). | |||||
|
ARHG8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.053880 (rank : 134) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z206, Q8C4I0, Q9Z1L7 | Gene names | Net1, Arhgef8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroepithelial cell-transforming gene 1 protein (Rho guanine nucleotide exchange factor 8). | |||||
|
ARHG9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.143585 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
ARHG9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.141780 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.174425 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHGA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.173979 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C033, Q5DU38, Q80VH8, Q8BW76, Q922S7 | Gene names | Arhgef10, Kiaa0294 | |||
|
Domain Architecture |
|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.111381 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.105131 (rank : 95) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.103521 (rank : 98) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.159484 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
ARHGG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.166582 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
ARHGG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.156421 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
BCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.141593 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CJ064_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.103389 (rank : 101) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS81, Q8N4A3 | Gene names | C10orf64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf64. | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050194 (rank : 136) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
DNMBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.074390 (rank : 121) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
DNMBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.077252 (rank : 119) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
DUET_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.061460 (rank : 127) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
ECT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.207485 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ECT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.212920 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.135941 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.145083 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.166260 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.104612 (rank : 96) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.103958 (rank : 97) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.108481 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.106517 (rank : 94) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
LBC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.124801 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |
|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
LRBA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.050076 (rank : 138) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
LYST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053095 (rank : 135) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
LYST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.055794 (rank : 133) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
MCF2L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.144524 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
MCF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.148759 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10911, P14919, Q5JYJ4, Q8IUF3, Q8IUF4, Q9UJB3 | Gene names | MCF2, DBL | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene DBL (Proto-oncogene MCF-2) [Contains: MCF2-transforming protein; DBL-transforming protein]. | |||||
|
NBEAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.061319 (rank : 128) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
NBEA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.050117 (rank : 137) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
NGEF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.164380 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
NGEF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.161456 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.060348 (rank : 130) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.060467 (rank : 129) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.081031 (rank : 115) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.109943 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.107254 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.191440 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.149077 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
RFFL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.086622 (rank : 109) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.077890 (rank : 118) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
RNF34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.092312 (rank : 106) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
RUFY3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.119725 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
RUFY3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.120754 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.071772 (rank : 122) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.064195 (rank : 125) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SOS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.080902 (rank : 116) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.116990 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.113303 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
VAV2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.083816 (rank : 113) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.086608 (rank : 110) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.086123 (rank : 111) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.085492 (rank : 112) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.092649 (rank : 105) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
VAV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.089904 (rank : 107) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
ZFY27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.247754 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5T4F4, Q5T4F1, Q5T4F2, Q5T4F3, Q8N1K0, Q8N6D6, Q8NCA0, Q8NDE4, Q96M08 | Gene names | ZFYVE27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
ZFY27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.248883 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
PKHF1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.86219e-138 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
PKHF1_HUMAN
|
||||||
| NC score | 0.998644 (rank : 2) | θ value | 3.4095e-124 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
PKHF2_HUMAN
|
||||||
| NC score | 0.980969 (rank : 3) | θ value | 2.92676e-75 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H8W4 | Gene names | PLEKHF2, ZFYVE18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2) (PH and FYVE domain-containing protein 2) (Phafin-2) (Zinc finger FYVE domain-containing protein 18). | |||||
|
PKHF2_MOUSE
|
||||||
| NC score | 0.980578 (rank : 4) | θ value | 2.24094e-75 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91WB4 | Gene names | Plekhf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.731069 (rank : 5) | θ value | 4.14116e-29 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.724888 (rank : 6) | θ value | 3.63628e-17 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.723896 (rank : 7) | θ value | 5.08577e-11 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.716356 (rank : 8) | θ value | 6.62687e-19 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.715782 (rank : 9) | θ value | 5.06226e-27 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
ZFY21_MOUSE
|
||||||
| NC score | 0.705676 (rank : 10) | θ value | 1.17247e-07 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY21_HUMAN
|
||||||
| NC score | 0.702290 (rank : 11) | θ value | 6.87365e-08 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.670830 (rank : 12) | θ value | 3.29651e-10 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
ZFY16_MOUSE
|
||||||
| NC score | 0.652996 (rank : 13) | θ value | 2.13673e-09 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
FGD2_MOUSE
|
||||||
| NC score | 0.643042 (rank : 14) | θ value | 3.07829e-16 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
ZFY16_HUMAN
|
||||||
| NC score | 0.642571 (rank : 15) | θ value | 2.13673e-09 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
FGD2_HUMAN
|
||||||
| NC score | 0.638950 (rank : 16) | θ value | 3.40345e-15 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.619085 (rank : 17) | θ value | 1.52774e-15 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.613406 (rank : 18) | θ value | 3.76295e-14 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.604669 (rank : 19) | θ value | 6.00763e-12 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.601076 (rank : 20) | θ value | 3.89403e-11 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
ZFYV1_HUMAN
|
||||||
| NC score | 0.559712 (rank : 21) | θ value | 8.40245e-06 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.556525 (rank : 22) | θ value | 1.69304e-06 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
ZFYV1_MOUSE
|
||||||
| NC score | 0.554732 (rank : 23) | θ value | 4.1701e-05 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.533681 (rank : 24) | θ value | 4.0297e-08 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.531548 (rank : 25) | θ value | 5.26297e-08 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.514413 (rank : 26) | θ value | 0.000461057 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.486577 (rank : 27) | θ value | 0.00020696 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
RBNS5_MOUSE
|
||||||
| NC score | 0.480288 (rank : 28) | θ value | 0.00020696 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.470146 (rank : 29) | θ value | 8.40245e-06 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.469369 (rank : 30) | θ value | 8.40245e-06 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
WDFY1_HUMAN
|
||||||
| NC score | 0.417867 (rank : 31) | θ value | 1.87187e-05 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IWB7, Q9H9D5, Q9P2B3 | Gene names | WDFY1, KIAA1435, WDF1, ZFYVE17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 1 (WD40- and FYVE domain- containing protein 1) (Phosphoinositide-binding protein 1) (FENS-1) (Zinc finger FYVE domain-containing protein 17). | |||||
|
WDFY3_MOUSE
|
||||||
| NC score | 0.409661 (rank : 32) | θ value | 1.47974e-10 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
ZFY19_HUMAN
|
||||||
| NC score | 0.408506 (rank : 33) | θ value | 0.279714 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
WDFY3_HUMAN
|
||||||
| NC score | 0.408492 (rank : 34) | θ value | 1.47974e-10 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
ZFY19_MOUSE
|
||||||
| NC score | 0.402186 (rank : 35) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DAZ9, Q8VCV7 | Gene names | Zfyve19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19. | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.398531 (rank : 36) | θ value | 8.11959e-09 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.396840 (rank : 37) | θ value | 4.0297e-08 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
WDFY2_MOUSE
|
||||||
| NC score | 0.396321 (rank : 38) | θ value | 5.44631e-05 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BUB4 | Gene names | Wdfy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2). | |||||
|
WDFY2_HUMAN
|
||||||
| NC score | 0.392718 (rank : 39) | θ value | 5.44631e-05 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96P53, Q96CS1 | Gene names | WDFY2, WDF2, ZFYVE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2) (Zinc finger FYVE domain-containing protein 22). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.391388 (rank : 40) | θ value | 2.36244e-08 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.389431 (rank : 41) | θ value | 7.59969e-07 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.317447 (rank : 42) | θ value | 4.0297e-08 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.316162 (rank : 43) | θ value | 1.06045e-08 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.315443 (rank : 44) | θ value | 8.97725e-08 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.309017 (rank : 45) | θ value | 4.92598e-06 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.269172 (rank : 46) | θ value | 7.59969e-07 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.268772 (rank : 47) | θ value | 1.69304e-06 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.257899 (rank : 48) | θ value | 1.43324e-05 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
PREX1_HUMAN
|
||||||
| NC score | 0.251027 (rank : 49) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
ZFY27_MOUSE
|
||||||
| NC score | 0.248883 (rank : 50) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
ZFY27_HUMAN
|
||||||
| NC score | 0.247754 (rank : 51) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5T4F4, Q5T4F1, Q5T4F2, Q5T4F3, Q8N1K0, Q8N6D6, Q8NCA0, Q8NDE4, Q96M08 | Gene names | ZFYVE27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.246798 (rank : 52) | θ value | 8.67504e-11 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.229848 (rank : 53) | θ value | 0.00228821 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.228319 (rank : 54) | θ value | 1.133e-10 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
ECT2_MOUSE
|
||||||
| NC score | 0.212920 (rank : 55) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ECT2_HUMAN
|
||||||
| NC score | 0.207485 (rank : 56) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.198089 (rank : 57) | θ value | 0.000158464 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.191440 (rank : 58) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.186109 (rank : 59) | θ value | 0.000786445 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ARHG5_HUMAN
|
||||||
| NC score | 0.180570 (rank : 60) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.174425 (rank : 61) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHGA_MOUSE
|
||||||
| NC score | 0.173979 (rank : 62) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C033, Q5DU38, Q80VH8, Q8BW76, Q922S7 | Gene names | Arhgef10, Kiaa0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHGG_HUMAN
|
||||||
| NC score | 0.166582 (rank : 63) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.166260 (rank : 64) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
NGEF_HUMAN
|
||||||
| NC score | 0.164380 (rank : 65) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
NGEF_MOUSE
|
||||||
| NC score | 0.161456 (rank : 66) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.159484 (rank : 67) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
ARHG2_MOUSE
|
||||||
| NC score | 0.156732 (rank : 68) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60875, O09115 | Gene names | Arhgef2, Lbcl1, Lfc | |||
|
Domain Architecture |

|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (Lymphoid blast crisis- like 1) (LBC'S first cousin) (Oncogene LFC) (RHOBIN). | |||||
|
ARHGG_MOUSE
|
||||||
| NC score | 0.156421 (rank : 69) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
ARHG4_HUMAN
|
||||||
| NC score | 0.155659 (rank : 70) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG4_MOUSE
|
||||||
| NC score | 0.155247 (rank : 71) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.149077 (rank : 72) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
MCF2_HUMAN
|
||||||
| NC score | 0.148759 (rank : 73) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10911, P14919, Q5JYJ4, Q8IUF3, Q8IUF4, Q9UJB3 | Gene names | MCF2, DBL | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene DBL (Proto-oncogene MCF-2) [Contains: MCF2-transforming protein; DBL-transforming protein]. | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.145083 (rank : 74) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
MCF2L_MOUSE
|
||||||
| NC score | 0.144524 (rank : 75) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
ARHG9_HUMAN
|
||||||
| NC score | 0.143585 (rank : 76) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
ARHG9_MOUSE
|
||||||
| NC score | 0.141780 (rank : 77) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.141593 (rank : 78) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.140846 (rank : 79) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.135941 (rank : 80) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
LBC_HUMAN
|
||||||
| NC score | 0.124801 (rank : 81) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |

|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
RUFY3_MOUSE
|
||||||
| NC score | 0.120754 (rank : 82) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
RUFY3_HUMAN
|
||||||
| NC score | 0.119725 (rank : 83) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.118857 (rank : 84) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
ARHG1_HUMAN
|
||||||
| NC score | 0.118855 (rank : 85) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.117378 (rank : 86) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.116990 (rank : 87) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.113303 (rank : 88) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.111381 (rank : 89) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.109943 (rank : 90) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.108481 (rank : 91) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.107254 (rank : 92) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
ARHG2_HUMAN
|
||||||
| NC score | 0.107042 (rank : 93) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92974, O75142, Q15079 | Gene names | ARHGEF2, KIAA0651, LFP40 | |||
|
Domain Architecture |

|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (GEF-H1 protein) (Proliferating cell nucleolar antigen p40). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.106517 (rank : 94) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.105131 (rank : 95) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.104612 (rank : 96) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.103958 (rank : 97) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.103521 (rank : 98) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
RFFL_MOUSE
|
||||||
| NC score | 0.103482 (rank : 99) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
RNF34_MOUSE
|
||||||
| NC score | 0.103390 (rank : 100) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
CJ064_HUMAN
|
||||||
| NC score | 0.103389 (rank : 101) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS81, Q8N4A3 | Gene names | C10orf64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf64. | |||||
|
ARHG8_HUMAN
|
||||||
| NC score | 0.101748 (rank : 102) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z628, Q12773, Q96D82, Q99903, Q9UEN6 | Gene names | NET1, ARHGEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroepithelial cell-transforming gene 1 protein (p65 Net1 proto- oncogene) (Rho guanine nucleotide exchange factor 8). | |||||
|
ARHG7_MOUSE
|
||||||
| NC score | 0.097118 (rank : 103) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.093608 (rank : 104) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
VAV_HUMAN
|
||||||
| NC score | 0.092649 (rank : 105) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
RNF34_HUMAN
|
||||||
| NC score | 0.092312 (rank : 106) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
VAV_MOUSE
|
||||||
| NC score | 0.089904 (rank : 107) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.088668 (rank : 108) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
RFFL_HUMAN
|
||||||
| NC score | 0.086622 (rank : 109) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
VAV2_MOUSE
|
||||||
| NC score | 0.086608 (rank : 110) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.086123 (rank : 111) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.085492 (rank : 112) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV2_HUMAN
|
||||||
| NC score | 0.083816 (rank : 113) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
ARHG3_MOUSE
|
||||||
| NC score | 0.081990 (rank : 114) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91X46, Q8CDM0, Q91VY4, Q99K14, Q9DC31 | Gene names | Arhgef3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3. | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.081031 (rank : 115) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.080902 (rank : 116) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
ARHG3_HUMAN
|
||||||
| NC score | 0.080706 (rank : 117) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.077890 (rank : 118) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
DNMBP_MOUSE
|
||||||
| NC score | 0.077252 (rank : 119) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
ARHG6_HUMAN
|
||||||
| NC score | 0.076356 (rank : 120) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
DNMBP_HUMAN
|
||||||
| NC score | 0.074390 (rank : 121) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.071772 (rank : 122) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
ARHG6_MOUSE
|
||||||
| NC score | 0.069305 (rank : 123) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.065003 (rank : 124) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.064195 (rank : 125) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.063784 (rank : 126) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
DUET_HUMAN
|
||||||
| NC score | 0.061460 (rank : 127) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
NBEAL_HUMAN
|
||||||
| NC score | 0.061319 (rank : 128) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.060467 (rank : 129) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.060348 (rank : 130) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.059437 (rank : 131) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.058109 (rank : 132) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.055794 (rank : 133) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
ARHG8_MOUSE
|
||||||
| NC score | 0.053880 (rank : 134) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z206, Q8C4I0, Q9Z1L7 | Gene names | Net1, Arhgef8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroepithelial cell-transforming gene 1 protein (Rho guanine nucleotide exchange factor 8). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.053095 (rank : 135) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.050194 (rank : 136) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
NBEA_MOUSE
|
||||||
| NC score | 0.050117 (rank : 137) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
LRBA_HUMAN
|
||||||
| NC score | 0.050076 (rank : 138) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.040609 (rank : 139) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
EXOC8_MOUSE
|
||||||
| NC score | 0.040419 (rank : 140) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGF7 | Gene names | Exoc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 8 (Exocyst complex 84 kDa subunit). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.039897 (rank : 141) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
S4A7_MOUSE
|
||||||
| NC score | 0.035880 (rank : 142) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BTY2, Q3U5H7 | Gene names | Slc4a7, Nbc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium bicarbonate cotransporter 3 (Solute carrier family 4 member 7). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.035850 (rank : 143) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.035011 (rank : 144) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
S4A7_HUMAN
|
||||||
| NC score | 0.034365 (rank : 145) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6M7, O60350, Q6AHZ9, Q9HC88, Q9UIB9 | Gene names | SLC4A7, BT, NBC2, NBC2B, NBC3, SBC2, SLC4A6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium bicarbonate cotransporter 3 (Sodium bicarbonate cotransporter 2) (Sodium bicarbonate cotransporter 2b) (Bicarbonate transporter) (Solute carrier family 4 member 7). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.030733 (rank : 146) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
S4A10_MOUSE
|
||||||
| NC score | 0.029103 (rank : 147) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DTL9, Q8CFS3, Q9EST0 | Gene names | Slc4a10, Kiaa4136, Ncbe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-driven chloride bicarbonate exchanger (Solute carrier family 4 member 10). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.029061 (rank : 148) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
S4A10_HUMAN
|
||||||
| NC score | 0.028927 (rank : 149) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6U841, Q4ZFX6, Q8TCP2, Q9HCQ6 | Gene names | SLC4A10, NCBE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-driven chloride bicarbonate exchanger (Solute carrier family 4 member 10). | |||||
|
ZDH21_MOUSE
|
||||||
| NC score | 0.020118 (rank : 150) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D270, Q6EMK1, Q80XQ3 | Gene names | Zdhhc21, Gramp3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21) (GABA-A receptor-associated membrane protein 3). | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.016551 (rank : 151) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.015039 (rank : 152) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.014206 (rank : 153) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.013542 (rank : 154) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.006196 (rank : 155) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
TRI14_MOUSE
|
||||||
| NC score | 0.003353 (rank : 156) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVW3, Q6A0C3, Q762I6, Q9D3G8 | Gene names | Trim14, Kiaa0129, Pub | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 14 (PU.1-binding protein). | |||||