Please be patient as the page loads
|
JHD2B_MOUSE
|
||||||
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
JHD2A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.964883 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
JHD2A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.961143 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.982204 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.920536 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.928153 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 7) | NC score | 0.770438 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 8) | NC score | 0.763630 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.082173 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SALL1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.007697 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
MFRP_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.022651 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.056243 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.050590 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.075892 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
UB2J1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.036875 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y385, Q5W0N4, Q9BZ32, Q9NQL3, Q9NY66, Q9P011, Q9P0S0, Q9UF10 | Gene names | UBE2J1, NCUBE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1) (Yeast ubiquitin-conjugating enzyme UBC6 homolog E) (HSUBC6e). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.049791 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.029186 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
INSM1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.019611 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.029988 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.043999 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.043979 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
TGBR3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.037140 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.015870 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.026919 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
UBP2L_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.034836 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
ADCY3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.016832 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
HXD11_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.008577 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31277, Q9NS02 | Gene names | HOXD11, HOX4F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4F). | |||||
|
MCR_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.015061 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
PKHF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.041583 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.049478 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
FANCJ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.022298 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX63, Q8NCI5 | Gene names | BRIP1, BACH1, FANCJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
PKHF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.040609 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.022763 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.012299 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.017178 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.024238 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
PARM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.032344 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.007413 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.014844 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.005800 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.011758 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.000991 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.019767 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.027293 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LAP2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.027835 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
PO4F2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.011247 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12837, Q13883, Q14987 | Gene names | POU4F2, BRN3B | |||
|
Domain Architecture |
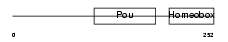
|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B). | |||||
|
PO4F2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.011222 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63934, Q63954 | Gene names | Pou4f2, Brn3b | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B) (Brn-3.2). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.015331 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
BRDT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.009040 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.011356 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
GLI3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.005953 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.014202 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.036348 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
NU153_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.029287 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PDE11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.006320 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PEX19_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.019595 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCI5, Q8CEE1, Q921H0, Q9CZC1, Q9QUQ1 | Gene names | Pex19, Pxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (PxF). | |||||
|
SQSTM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.017799 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
ARAF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | -0.001089 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.036149 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.018956 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.004690 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
DVL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.007852 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |
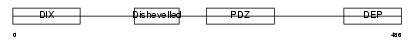
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.010879 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
HXA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.004530 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.005204 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.002263 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.012222 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
REPS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.011768 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.016981 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
STON2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.028287 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
ABCAD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.004123 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
APC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.047012 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.021787 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.003829 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.011868 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.014788 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.016837 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.015259 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RBGPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.011679 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2M9, O75872, Q9HAB0, Q9UFJ7, Q9UQ15 | Gene names | RAB3GAP2, KIAA0839 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein non-catalytic subunit (Rab3 GTPase- activating protein 150 kDa subunit) (Rab3-GAP p150) (Rab3-GAP regulatory subunit) (RAB3-GAP150) (RGAP-iso). | |||||
|
RUSC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.009245 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
SPAG8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.021526 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99932, Q12937, Q5TCV8, Q8WWB4 | Gene names | SPAG8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 8 (Sperm membrane protein 1) (SMP-1) (HSD-1) (Sperm membrane protein BS-84). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.031174 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.036209 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.003434 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
AF17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.008190 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.011558 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.002990 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.003229 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.010028 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.017407 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.010732 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.017819 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.000445 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3SY56 | Gene names | SP6, KLF14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.008575 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TR150_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.011262 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.017147 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UB2J1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.020672 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJZ4, Q9DC92 | Gene names | Ube2j1, Ncube1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1). | |||||
|
UBP2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.025375 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |
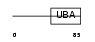
|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
XYLT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.009374 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811B1, Q9EPL1 | Gene names | Xylt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (Peptide O- xylosyltransferase 1). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.001365 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.025209 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ADRM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.014629 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |
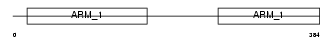
|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
ADRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.015430 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.012248 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.005294 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.003590 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.020627 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.014542 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.007492 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PDK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.004611 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16654 | Gene names | PDK4 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 4, mitochondrial precursor (EC 2.7.11.2) (Pyruvate dehydrogenase kinase isoform 4). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.011350 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PKHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.009492 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TCZ9 | Gene names | PKHD1, FCYT, TIGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney and hepatic disease 1 precursor (Fibrocystin) (Polyductin) (Tigmin). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.007170 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
SYMPK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.012877 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.009442 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.982204 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.964883 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
JHD2A_MOUSE
|
||||||
| NC score | 0.961143 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.928153 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.920536 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.770438 (rank : 7) | θ value | 3.48949e-44 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.763630 (rank : 8) | θ value | 2.50074e-42 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.082173 (rank : 9) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.075892 (rank : 10) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.056243 (rank : 11) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.050590 (rank : 12) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.049791 (rank : 13) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.049478 (rank : 14) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.047012 (rank : 15) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.043999 (rank : 16) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.043979 (rank : 17) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
PKHF1_HUMAN
|
||||||
| NC score | 0.041583 (rank : 18) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
PKHF1_MOUSE
|
||||||
| NC score | 0.040609 (rank : 19) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
TGBR3_HUMAN
|
||||||
| NC score | 0.037140 (rank : 20) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
UB2J1_HUMAN
|
||||||
| NC score | 0.036875 (rank : 21) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y385, Q5W0N4, Q9BZ32, Q9NQL3, Q9NY66, Q9P011, Q9P0S0, Q9UF10 | Gene names | UBE2J1, NCUBE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1) (Yeast ubiquitin-conjugating enzyme UBC6 homolog E) (HSUBC6e). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.036348 (rank : 22) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.036209 (rank : 23) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.036149 (rank : 24) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
UBP2L_MOUSE
|
||||||
| NC score | 0.034836 (rank : 25) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
PARM1_HUMAN
|
||||||
| NC score | 0.032344 (rank : 26) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.031174 (rank : 27) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.029988 (rank : 28) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.029287 (rank : 29) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.029186 (rank : 30) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.028287 (rank : 31) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
LAP2A_HUMAN
|
||||||
| NC score | 0.027835 (rank : 32) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.027293 (rank : 33) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.026919 (rank : 34) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
UBP2L_HUMAN
|
||||||
| NC score | 0.025375 (rank : 35) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.025209 (rank : 36) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.024238 (rank : 37) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.022763 (rank : 38) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.022651 (rank : 39) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
FANCJ_HUMAN
|
||||||
| NC score | 0.022298 (rank : 40) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX63, Q8NCI5 | Gene names | BRIP1, BACH1, FANCJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.021787 (rank : 41) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
SPAG8_HUMAN
|
||||||
| NC score | 0.021526 (rank : 42) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99932, Q12937, Q5TCV8, Q8WWB4 | Gene names | SPAG8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 8 (Sperm membrane protein 1) (SMP-1) (HSD-1) (Sperm membrane protein BS-84). | |||||
|
UB2J1_MOUSE
|
||||||
| NC score | 0.020672 (rank : 43) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJZ4, Q9DC92 | Gene names | Ube2j1, Ncube1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.020627 (rank : 44) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.019767 (rank : 45) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
INSM1_MOUSE
|
||||||
| NC score | 0.019611 (rank : 46) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
PEX19_MOUSE
|
||||||
| NC score | 0.019595 (rank : 47) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCI5, Q8CEE1, Q921H0, Q9CZC1, Q9QUQ1 | Gene names | Pex19, Pxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (PxF). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.018956 (rank : 48) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.017819 (rank : 49) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SQSTM_HUMAN
|
||||||
| NC score | 0.017799 (rank : 50) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.017407 (rank : 51) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.017178 (rank : 52) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.017147 (rank : 53) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.016981 (rank : 54) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.016837 (rank : 55) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ADCY3_HUMAN
|
||||||
| NC score | 0.016832 (rank : 56) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.015870 (rank : 57) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
ADRM1_MOUSE
|
||||||
| NC score | 0.015430 (rank : 58) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.015331 (rank : 59) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.015259 (rank : 60) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
MCR_MOUSE
|
||||||
| NC score | 0.015061 (rank : 61) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.014844 (rank : 62) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.014788 (rank : 63) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
ADRM1_HUMAN
|
||||||
| NC score | 0.014629 (rank : 64) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.014542 (rank : 65) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.014202 (rank : 66) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SYMPK_MOUSE
|
||||||
| NC score | 0.012877 (rank : 67) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.012299 (rank : 68) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.012248 (rank : 69) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.012222 (rank : 70) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.011868 (rank : 71) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
REPS1_HUMAN
|
||||||
| NC score | 0.011768 (rank : 72) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.011758 (rank : 73) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
RBGPR_HUMAN
|
||||||
| NC score | 0.011679 (rank : 74) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2M9, O75872, Q9HAB0, Q9UFJ7, Q9UQ15 | Gene names | RAB3GAP2, KIAA0839 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein non-catalytic subunit (Rab3 GTPase- activating protein 150 kDa subunit) (Rab3-GAP p150) (Rab3-GAP regulatory subunit) (RAB3-GAP150) (RGAP-iso). | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.011558 (rank : 75) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.011356 (rank : 76) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.011350 (rank : 77) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.011262 (rank : 78) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
PO4F2_HUMAN
|
||||||
| NC score | 0.011247 (rank : 79) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12837, Q13883, Q14987 | Gene names | POU4F2, BRN3B | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B). | |||||
|
PO4F2_MOUSE
|
||||||
| NC score | 0.011222 (rank : 80) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63934, Q63954 | Gene names | Pou4f2, Brn3b | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B) (Brn-3.2). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.010879 (rank : 81) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.010732 (rank : 82) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.010028 (rank : 83) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
PKHD1_HUMAN
|
||||||
| NC score | 0.009492 (rank : 84) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TCZ9 | Gene names | PKHD1, FCYT, TIGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney and hepatic disease 1 precursor (Fibrocystin) (Polyductin) (Tigmin). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.009442 (rank : 85) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
XYLT1_MOUSE
|
||||||
| NC score | 0.009374 (rank : 86) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811B1, Q9EPL1 | Gene names | Xylt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (Peptide O- xylosyltransferase 1). | |||||
|
RUSC1_HUMAN
|
||||||
| NC score | 0.009245 (rank : 87) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.009040 (rank : 88) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
HXD11_HUMAN
|
||||||
| NC score | 0.008577 (rank : 89) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31277, Q9NS02 | Gene names | HOXD11, HOX4F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4F). | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.008575 (rank : 90) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.008190 (rank : 91) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
DVL1_MOUSE
|
||||||
| NC score | 0.007852 (rank : 92) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
SALL1_HUMAN
|
||||||
| NC score | 0.007697 (rank : 93) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.007492 (rank : 94) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.007413 (rank : 95) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.007170 (rank : 96) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PDE11_MOUSE
|
||||||
| NC score | 0.006320 (rank : 97) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
GLI3_HUMAN
|
||||||
| NC score | 0.005953 (rank : 98) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.005800 (rank : 99) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.005294 (rank : 100) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.005204 (rank : 101) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.004690 (rank : 102) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
PDK4_HUMAN
|
||||||
| NC score | 0.004611 (rank : 103) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16654 | Gene names | PDK4 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 4, mitochondrial precursor (EC 2.7.11.2) (Pyruvate dehydrogenase kinase isoform 4). | |||||
|
HXA5_HUMAN
|
||||||
| NC score | 0.004530 (rank : 104) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
ABCAD_MOUSE
|
||||||
| NC score | 0.004123 (rank : 105) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
GLI3_MOUSE
|
||||||
| NC score | 0.003829 (rank : 106) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.003590 (rank : 107) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.003434 (rank : 108) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.003229 (rank : 109) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.002990 (rank : 110) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.002263 (rank : 111) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.001365 (rank : 112) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.000991 (rank : 113) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
SP6_HUMAN
|
||||||
| NC score | 0.000445 (rank : 114) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3SY56 | Gene names | SP6, KLF14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14). | |||||
|
ARAF_MOUSE
|
||||||
| NC score | -0.001089 (rank : 115) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||