Please be patient as the page loads
|
HAIR_HUMAN
|
||||||
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HAIR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 172 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.961892 (rank : 2) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 4.84666e-62 (rank : 3) | NC score | 0.774607 (rank : 3) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 5.35859e-61 (rank : 4) | NC score | 0.767898 (rank : 5) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 8.00737e-57 (rank : 5) | NC score | 0.771658 (rank : 4) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 6) | NC score | 0.760947 (rank : 7) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 2.50074e-42 (rank : 7) | NC score | 0.763630 (rank : 6) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
JHD2A_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 8) | NC score | 0.758021 (rank : 8) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 9) | NC score | 0.075509 (rank : 15) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 10) | NC score | 0.075496 (rank : 16) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 11) | NC score | 0.053050 (rank : 45) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.067080 (rank : 25) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.038733 (rank : 84) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.048052 (rank : 58) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.067825 (rank : 23) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.024404 (rank : 116) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PTDSR_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.109396 (rank : 9) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC1, Q86VY0, Q8IUM5, Q9Y4E2 | Gene names | PTDSR, KIAA0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor). | |||||
|
PTDSR_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.109150 (rank : 10) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERI5, Q80TX1 | Gene names | Ptdsr, Kiaa0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor) (Apoptotic cell clearance receptor PtdSerR). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.018659 (rank : 135) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.080827 (rank : 13) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.039442 (rank : 81) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.038831 (rank : 83) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.043595 (rank : 63) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.009129 (rank : 164) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.069441 (rank : 22) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
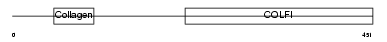
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.051127 (rank : 52) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.054149 (rank : 41) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.034206 (rank : 96) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.059606 (rank : 30) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
CN004_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.073698 (rank : 18) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CN004_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.072399 (rank : 19) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.060226 (rank : 29) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.052358 (rank : 47) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
K1849_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.035070 (rank : 95) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.029614 (rank : 105) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.054300 (rank : 39) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.059528 (rank : 31) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.039473 (rank : 80) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.040249 (rank : 78) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.074763 (rank : 17) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.038624 (rank : 85) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.030967 (rank : 99) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
ZN444_HUMAN
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.003293 (rank : 181) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N0Y2, Q8TEQ9, Q8WU35, Q9NUU1 | Gene names | ZNF444, EZF2, ZSCAN17 | |||
|
Domain Architecture |
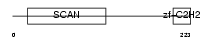
|
|||||
| Description | Zinc finger protein 444 (Endothelial zinc finger protein 2) (EZF-2) (Zinc finger and SCAN domain-containing protein 17). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.041269 (rank : 74) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.011722 (rank : 159) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.004239 (rank : 179) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
PRR13_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.087656 (rank : 12) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.038406 (rank : 87) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ZN414_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.026316 (rank : 112) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.030274 (rank : 101) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
CF150_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.042442 (rank : 69) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
CM018_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.042808 (rank : 67) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H714, Q5W051, Q6PJ74, Q6PK94, Q86XH7, Q8N5J6 | Gene names | C13orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf18. | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.062252 (rank : 27) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.026961 (rank : 108) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.030749 (rank : 100) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.043769 (rank : 62) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.041758 (rank : 71) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RXRB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.010155 (rank : 161) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P28702, P28703 | Gene names | RXRB, NR2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.035866 (rank : 93) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
CO8A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.043910 (rank : 61) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
CO9A2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.048860 (rank : 57) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
COJA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.051866 (rank : 51) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.026017 (rank : 113) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.017470 (rank : 140) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.038233 (rank : 88) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.038557 (rank : 86) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.070163 (rank : 21) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CDX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.007185 (rank : 172) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |
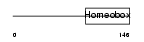
|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
DPOG2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.026833 (rank : 111) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHN1, O00419, Q9UK35, Q9UK94 | Gene names | POLG2, MTPOLB | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit gamma 2, mitochondrial precursor (EC 2.7.7.7) (Mitochondrial DNA polymerase accessory subunit) (PolG-beta) (MtPolB) (DNA polymerase gamma accessory 55 kDa subunit) (p55). | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.022975 (rank : 121) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.037844 (rank : 90) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.017664 (rank : 138) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.019874 (rank : 131) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.032988 (rank : 97) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.020770 (rank : 129) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.043042 (rank : 66) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.037109 (rank : 92) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.070662 (rank : 20) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.058325 (rank : 34) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
HKR3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.001183 (rank : 185) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10074 | Gene names | HKR3 | |||
|
Domain Architecture |
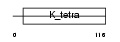
|
|||||
| Description | Krueppel-related zinc finger protein 3 (Protein HKR3). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.022137 (rank : 124) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.026850 (rank : 109) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.026843 (rank : 110) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.040272 (rank : 77) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.016287 (rank : 143) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.019752 (rank : 132) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.040775 (rank : 76) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.007688 (rank : 171) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
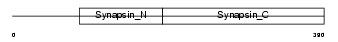
SYN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.017823 (rank : 137) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |
|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
A2MG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.024590 (rank : 115) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61838, Q60628 | Gene names | A2m | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M) [Contains: Alpha-2- macroglobulin 165 kDa subunit; Alpha-2-macroglobulin 35 kDa subunit]. | |||||
|
CBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.017556 (rank : 139) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CI047_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.043459 (rank : 64) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZRZ4, Q5SQD7, Q7Z568, Q8N1V4 | Gene names | C9orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf47 precursor. | |||||
|
CO8A2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.040995 (rank : 75) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
GDPD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.014209 (rank : 151) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCC8, Q9NXJ6 | Gene names | GDPD2, GDE3, OBDPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 2 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 3) (Osteoblast differentiation promoting factor). | |||||
|
NKX22_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.005520 (rank : 176) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95096 | Gene names | NKX2-2, NKX2.2, NKX2B | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.2 (Homeobox protein NK-2 homolog B). | |||||
|
PDLI2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.013265 (rank : 153) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
PRP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.063837 (rank : 26) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
RAVR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.021468 (rank : 125) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY67, Q8IY60, Q8TF24 | Gene names | RAVER1, KIAA1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
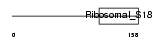
RT18B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.023885 (rank : 119) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N84, Q3TLA9, Q9CRK0, Q9DCR8 | Gene names | Mrps18b | |||
|
Domain Architecture |
|
|||||
| Description | 28S ribosomal protein S18b, mitochondrial precursor (MRP-S18-b) (Mrps18b) (MRP-S18-2). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.008901 (rank : 166) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
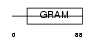
WBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.029743 (rank : 104) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969T9, O95638 | Gene names | WBP2 | |||
|
Domain Architecture |
|
|||||
| Description | WW domain-binding protein 2 (WBP-2). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.035692 (rank : 94) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.014526 (rank : 149) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.043194 (rank : 65) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.055766 (rank : 37) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CO6A2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.041388 (rank : 73) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
FBX38_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.015752 (rank : 145) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
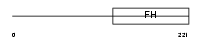
FOXD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.005091 (rank : 177) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35392 | Gene names | Foxd2, Mf2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D2 (Mesoderm/mesenchyme forkhead 2) (MF-2). | |||||
|
KIT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.000573 (rank : 186) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05532, Q61415, Q61416, Q61417, Q6LEE9 | Gene names | Kit, Sl | |||
|
Domain Architecture |

|
|||||
| Description | Mast/stem cell growth factor receptor precursor (EC 2.7.10.1) (SCFR) (Proto-oncogene tyrosine-protein kinase Kit) (c-kit) (CD117 antigen). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.053480 (rank : 44) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.008342 (rank : 167) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.014691 (rank : 147) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.001466 (rank : 184) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.039909 (rank : 79) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.024639 (rank : 114) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.037935 (rank : 89) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.029879 (rank : 102) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.045122 (rank : 59) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.012248 (rank : 155) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CK042_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.024224 (rank : 117) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5U0 | Gene names | C11orf42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf42. | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.019476 (rank : 133) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
DAB2P_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.009115 (rank : 165) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.004390 (rank : 178) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.002191 (rank : 183) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
PCX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.021145 (rank : 127) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.002452 (rank : 182) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ROS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.000019 (rank : 187) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 907 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08922, Q15368 | Gene names | ROS1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ROS precursor (EC 2.7.10.1) (c- ros-1). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.022914 (rank : 122) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.009750 (rank : 162) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.006153 (rank : 175) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
CD19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.021252 (rank : 126) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |
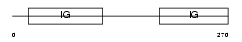
|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.017189 (rank : 141) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
CN032_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.042765 (rank : 68) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CO4A4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.052199 (rank : 49) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
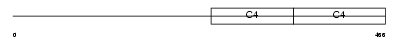
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.061538 (rank : 28) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.037740 (rank : 91) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
COGA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.044989 (rank : 60) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.041545 (rank : 72) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.042330 (rank : 70) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
IRX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.011886 (rank : 157) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |
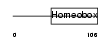
|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.004119 (rank : 180) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.006658 (rank : 173) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.029779 (rank : 103) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
RAVR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.020999 (rank : 128) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.014219 (rank : 150) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.020534 (rank : 130) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.018918 (rank : 134) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
VCY1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.106009 (rank : 11) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.011575 (rank : 160) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
AP180_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.014594 (rank : 148) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.027708 (rank : 106) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.053703 (rank : 43) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CU030_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.027579 (rank : 107) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UFM2 | Gene names | C21orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C21orf30. | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.013927 (rank : 152) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.016369 (rank : 142) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
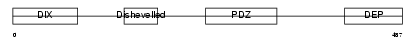
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.016098 (rank : 144) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.007992 (rank : 169) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.024217 (rank : 118) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.014840 (rank : 146) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.011900 (rank : 156) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.011753 (rank : 158) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.022464 (rank : 123) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.012809 (rank : 154) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.039104 (rank : 82) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.032828 (rank : 98) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MIS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.009235 (rank : 163) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
NUP37_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.007728 (rank : 170) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFH4, Q9H644 | Gene names | NUP37 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleoporin Nup37 (p37). | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | -0.001364 (rank : 188) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
TNR11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.006440 (rank : 174) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.008334 (rank : 168) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
TULP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.017870 (rank : 136) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
WBP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.023150 (rank : 120) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97765 | Gene names | Wbp2 | |||
|
Domain Architecture |
|
|||||
| Description | WW domain-binding protein 2 (WBP-2). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.054154 (rank : 40) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO1A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.054120 (rank : 42) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.058651 (rank : 33) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.051925 (rank : 50) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.054733 (rank : 38) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.050562 (rank : 54) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.050254 (rank : 55) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050781 (rank : 53) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.050241 (rank : 56) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.067711 (rank : 24) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.075860 (rank : 14) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.058165 (rank : 35) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.056218 (rank : 36) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SC24A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.052646 (rank : 46) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.052351 (rank : 48) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.058675 (rank : 32) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 172 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.961892 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.774607 (rank : 3) | θ value | 4.84666e-62 (rank : 3) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.771658 (rank : 4) | θ value | 8.00737e-57 (rank : 5) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.767898 (rank : 5) | θ value | 5.35859e-61 (rank : 4) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.763630 (rank : 6) | θ value | 2.50074e-42 (rank : 7) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.760947 (rank : 7) | θ value | 1.32601e-43 (rank : 6) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2A_MOUSE
|
||||||
| NC score | 0.758021 (rank : 8) | θ value | 1.62093e-41 (rank : 8) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
PTDSR_HUMAN
|
||||||
| NC score | 0.109396 (rank : 9) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC1, Q86VY0, Q8IUM5, Q9Y4E2 | Gene names | PTDSR, KIAA0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor). | |||||
|
PTDSR_MOUSE
|
||||||
| NC score | 0.109150 (rank : 10) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERI5, Q80TX1 | Gene names | Ptdsr, Kiaa0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor) (Apoptotic cell clearance receptor PtdSerR). | |||||
|
VCY1_HUMAN
|
||||||
| NC score | 0.106009 (rank : 11) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.087656 (rank : 12) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.080827 (rank : 13) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.075860 (rank : 14) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.075509 (rank : 15) | θ value | 0.00020696 (rank : 9) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.075496 (rank : 16) | θ value | 0.00035302 (rank : 10) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.074763 (rank : 17) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.073698 (rank : 18) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.072399 (rank : 19) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.070662 (rank : 20) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.070163 (rank : 21) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.069441 (rank : 22) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.067825 (rank : 23) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.067711 (rank : 24) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.067080 (rank : 25) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.063837 (rank : 26) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.062252 (rank : 27) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.061538 (rank : 28) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.060226 (rank : 29) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.059606 (rank : 30) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.059528 (rank : 31) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.058675 (rank : 32) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.058651 (rank : 33) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.058325 (rank : 34) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.058165 (rank : 35) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.056218 (rank : 36) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.055766 (rank : 37) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.054733 (rank : 38) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.054300 (rank : 39) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.054154 (rank : 40) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.054149 (rank : 41) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CO1A2_HUMAN
|
||||||
| NC score | 0.054120 (rank : 42) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.053703 (rank : 43) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.053480 (rank : 44) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.053050 (rank : 45) | θ value | 0.000786445 (rank : 11) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.052646 (rank : 46) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.052358 (rank : 47) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.052351 (rank : 48) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.052199 (rank : 49) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.051925 (rank : 50) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.051866 (rank : 51) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.051127 (rank : 52) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.050781 (rank : 53) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.050562 (rank : 54) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.050254 (rank : 55) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.050241 (rank : 56) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO9A2_MOUSE
|
||||||
| NC score | 0.048860 (rank : 57) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.048052 (rank : 58) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.045122 (rank : 59) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.044989 (rank : 60) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
CO8A1_HUMAN
|
||||||
| NC score | 0.043910 (rank : 61) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.043769 (rank : 62) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.043595 (rank : 63) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
CI047_HUMAN
|
||||||
| NC score | 0.043459 (rank : 64) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZRZ4, Q5SQD7, Q7Z568, Q8N1V4 | Gene names | C9orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf47 precursor. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.043194 (rank : 65) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.043042 (rank : 66) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
CM018_HUMAN
|
||||||
| NC score | 0.042808 (rank : 67) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H714, Q5W051, Q6PJ74, Q6PK94, Q86XH7, Q8N5J6 | Gene names | C13orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf18. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.042765 (rank : 68) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CF150_HUMAN
|
||||||
| NC score | 0.042442 (rank : 69) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.042330 (rank : 70) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.041758 (rank : 71) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.041545 (rank : 72) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CO6A2_HUMAN
|
||||||
| NC score | 0.041388 (rank : 73) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.041269 (rank : 74) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CO8A2_MOUSE
|
||||||
| NC score | 0.040995 (rank : 75) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.040775 (rank : 76) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.040272 (rank : 77) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.040249 (rank : 78) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.039909 (rank : 79) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.039473 (rank : 80) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.039442 (rank : 81) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.039104 (rank : 82) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.038831 (rank : 83) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.038733 (rank : 84) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.038624 (rank : 85) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.038557 (rank : 86) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.038406 (rank : 87) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.038233 (rank : 88) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.037935 (rank : 89) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.037844 (rank : 90) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.037740 (rank : 91) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.037109 (rank : 92) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.035866 (rank : 93) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.035692 (rank : 94) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.035070 (rank : 95) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.034206 (rank : 96) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.032988 (rank : 97) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.032828 (rank : 98) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.030967 (rank : 99) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.030749 (rank : 100) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.030274 (rank : 101) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.029879 (rank : 102) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.029779 (rank : 103) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
WBP2_HUMAN
|
||||||
| NC score | 0.029743 (rank : 104) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969T9, O95638 | Gene names | WBP2 | |||
|
Domain Architecture |

|
|||||
| Description | WW domain-binding protein 2 (WBP-2). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.029614 (rank : 105) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.027708 (rank : 106) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CU030_HUMAN
|
||||||
| NC score | 0.027579 (rank : 107) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UFM2 | Gene names | C21orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C21orf30. | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.026961 (rank : 108) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.026850 (rank : 109) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.026843 (rank : 110) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
DPOG2_HUMAN
|
||||||
| NC score | 0.026833 (rank : 111) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHN1, O00419, Q9UK35, Q9UK94 | Gene names | POLG2, MTPOLB | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit gamma 2, mitochondrial precursor (EC 2.7.7.7) (Mitochondrial DNA polymerase accessory subunit) (PolG-beta) (MtPolB) (DNA polymerase gamma accessory 55 kDa subunit) (p55). | |||||
|
ZN414_HUMAN
|
||||||
| NC score | 0.026316 (rank : 112) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.026017 (rank : 113) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.024639 (rank : 114) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
A2MG_MOUSE
|
||||||
| NC score | 0.024590 (rank : 115) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61838, Q60628 | Gene names | A2m | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M) [Contains: Alpha-2- macroglobulin 165 kDa subunit; Alpha-2-macroglobulin 35 kDa subunit]. | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.024404 (rank : 116) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
CK042_HUMAN
|
||||||
| NC score | 0.024224 (rank : 117) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5U0 | Gene names | C11orf42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf42. | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.024217 (rank : 118) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
RT18B_MOUSE
|
||||||
| NC score | 0.023885 (rank : 119) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N84, Q3TLA9, Q9CRK0, Q9DCR8 | Gene names | Mrps18b | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S18b, mitochondrial precursor (MRP-S18-b) (Mrps18b) (MRP-S18-2). | |||||
|
WBP2_MOUSE
|
||||||
| NC score | 0.023150 (rank : 120) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97765 | Gene names | Wbp2 | |||
|
Domain Architecture |

|
|||||
| Description | WW domain-binding protein 2 (WBP-2). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.022975 (rank : 121) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.022914 (rank : 122) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.022464 (rank : 123) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.022137 (rank : 124) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
RAVR1_HUMAN
|
||||||
| NC score | 0.021468 (rank : 125) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY67, Q8IY60, Q8TF24 | Gene names | RAVER1, KIAA1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
CD19_HUMAN
|
||||||
| NC score | 0.021252 (rank : 126) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |

|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
PCX3_MOUSE
|
||||||
| NC score | 0.021145 (rank : 127) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
RAVR1_MOUSE
|
||||||
| NC score | 0.020999 (rank : 128) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.020770 (rank : 129) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.020534 (rank : 130) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.019874 (rank : 131) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.019752 (rank : 132) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.019476 (rank : 133) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.018918 (rank : 134) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.018659 (rank : 135) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
TULP1_MOUSE
|
||||||
| NC score | 0.017870 (rank : 136) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
SYN3_HUMAN
|
||||||
| NC score | 0.017823 (rank : 137) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.017664 (rank : 138) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.017556 (rank : 139) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.017470 (rank : 140) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.017189 (rank : 141) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.016369 (rank : 142) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.016287 (rank : 143) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.016098 (rank : 144) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
FBX38_HUMAN
|
||||||
| NC score | 0.015752 (rank : 145) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.014840 (rank : 146) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.014691 (rank : 147) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.014594 (rank : 148) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.014526 (rank : 149) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.014219 (rank : 150) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
GDPD2_HUMAN
|
||||||
| NC score | 0.014209 (rank : 151) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCC8, Q9NXJ6 | Gene names | GDPD2, GDE3, OBDPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 2 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 3) (Osteoblast differentiation promoting factor). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.013927 (rank : 152) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
PDLI2_HUMAN
|
||||||
| NC score | 0.013265 (rank : 153) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.012809 (rank : 154) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.012248 (rank : 155) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.011900 (rank : 156) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IRX5_HUMAN
|
||||||
| NC score | 0.011886 (rank : 157) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.011753 (rank : 158) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.011722 (rank : 159) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.011575 (rank : 160) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
RXRB_HUMAN
|
||||||
| NC score | 0.010155 (rank : 161) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P28702, P28703 | Gene names | RXRB, NR2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.009750 (rank : 162) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
MIS_HUMAN
|
||||||
| NC score | 0.009235 (rank : 163) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.009129 (rank : 164) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
DAB2P_MOUSE
|
||||||
| NC score | 0.009115 (rank : 165) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.008901 (rank : 166) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.008342 (rank : 167) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.008334 (rank : 168) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.007992 (rank : 169) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
NUP37_HUMAN
|
||||||
| NC score | 0.007728 (rank : 170) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFH4, Q9H644 | Gene names | NUP37 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin Nup37 (p37). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.007688 (rank : 171) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CDX1_HUMAN
|
||||||
| NC score | 0.007185 (rank : 172) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.006658 (rank : 173) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
TNR11_MOUSE
|
||||||
| NC score | 0.006440 (rank : 174) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.006153 (rank : 175) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
NKX22_HUMAN
|
||||||
| NC score | 0.005520 (rank : 176) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95096 | Gene names | NKX2-2, NKX2.2, NKX2B | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.2 (Homeobox protein NK-2 homolog B). | |||||
|
FOXD2_MOUSE
|
||||||
| NC score | 0.005091 (rank : 177) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35392 | Gene names | Foxd2, Mf2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D2 (Mesoderm/mesenchyme forkhead 2) (MF-2). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.004390 (rank : 178) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.004239 (rank : 179) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.004119 (rank : 180) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
ZN444_HUMAN
|
||||||
| NC score | 0.003293 (rank : 181) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N0Y2, Q8TEQ9, Q8WU35, Q9NUU1 | Gene names | ZNF444, EZF2, ZSCAN17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 444 (Endothelial zinc finger protein 2) (EZF-2) (Zinc finger and SCAN domain-containing protein 17). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.002452 (rank : 182) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.002191 (rank : 183) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.001466 (rank : 184) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
HKR3_HUMAN
|
||||||
| NC score | 0.001183 (rank : 185) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10074 | Gene names | HKR3 | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-related zinc finger protein 3 (Protein HKR3). | |||||
|
KIT_MOUSE
|
||||||
| NC score | 0.000573 (rank : 186) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05532, Q61415, Q61416, Q61417, Q6LEE9 | Gene names | Kit, Sl | |||
|
Domain Architecture |

|
|||||
| Description | Mast/stem cell growth factor receptor precursor (EC 2.7.10.1) (SCFR) (Proto-oncogene tyrosine-protein kinase Kit) (c-kit) (CD117 antigen). | |||||
|
ROS_HUMAN
|
||||||
| NC score | 0.000019 (rank : 187) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 907 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08922, Q15368 | Gene names | ROS1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ROS precursor (EC 2.7.10.1) (c- ros-1). | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | -0.001364 (rank : 188) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||