Please be patient as the page loads
|
RHG06_HUMAN
|
||||||
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RHG06_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994230 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 3) | NC score | 0.719414 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 4) | NC score | 0.799530 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 5) | NC score | 0.722793 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 6) | NC score | 0.723863 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 7) | NC score | 0.710916 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
RHG07_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 8) | NC score | 0.733934 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 9) | NC score | 0.785582 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 10) | NC score | 0.789867 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 11) | NC score | 0.783693 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 12) | NC score | 0.736290 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 13) | NC score | 0.809581 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 14) | NC score | 0.790769 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 15) | NC score | 0.723168 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
STAR8_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 16) | NC score | 0.701657 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
CHIO_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 17) | NC score | 0.746619 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 18) | NC score | 0.699747 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CHIO_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 19) | NC score | 0.715408 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
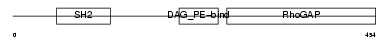
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 20) | NC score | 0.408678 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 21) | NC score | 0.413929 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 22) | NC score | 0.768869 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 23) | NC score | 0.692808 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 24) | NC score | 0.772893 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 25) | NC score | 0.715677 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 26) | NC score | 0.691475 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 27) | NC score | 0.710661 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 28) | NC score | 0.741170 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 29) | NC score | 0.720618 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 30) | NC score | 0.659217 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 31) | NC score | 0.693040 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 32) | NC score | 0.683730 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 33) | NC score | 0.779891 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 34) | NC score | 0.692204 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 35) | NC score | 0.777791 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 36) | NC score | 0.643785 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 37) | NC score | 0.688818 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 38) | NC score | 0.645732 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 39) | NC score | 0.598379 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 40) | NC score | 0.688082 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 41) | NC score | 0.733744 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG18_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 42) | NC score | 0.778129 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 43) | NC score | 0.690301 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 44) | NC score | 0.587288 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 45) | NC score | 0.768998 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 46) | NC score | 0.775499 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 47) | NC score | 0.731686 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 48) | NC score | 0.624449 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 49) | NC score | 0.746671 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 50) | NC score | 0.789746 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 51) | NC score | 0.514064 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 52) | NC score | 0.532979 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 53) | NC score | 0.718867 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 54) | NC score | 0.710540 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 55) | NC score | 0.717573 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 56) | NC score | 0.697146 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
K1688_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 57) | NC score | 0.543386 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 58) | NC score | 0.555386 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
OCRL_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 59) | NC score | 0.344616 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
P85A_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 60) | NC score | 0.350449 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
I5P2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 61) | NC score | 0.336942 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 62) | NC score | 0.046623 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 63) | NC score | 0.055497 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 64) | NC score | 0.335745 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 65) | NC score | 0.002109 (rank : 150) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 66) | NC score | 0.042726 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
DYDC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.073722 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.026699 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.018518 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.028548 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.014321 (rank : 128) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
K1219_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.030614 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
DNMBP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.044878 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.021182 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NECD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.009853 (rank : 138) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
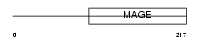
|
|||||
| Description | Necdin. | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.004823 (rank : 146) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.040775 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.023217 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
ZSCA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | -0.000613 (rank : 154) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBB4, Q6WLH8, Q86WS8 | Gene names | ZSCAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 1. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.022591 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.003165 (rank : 149) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
HMCS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.012118 (rank : 130) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8JZK9, Q3UXI4 | Gene names | Hmgcs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hydroxymethylglutaryl-CoA synthase, cytoplasmic (EC 2.3.3.10) (HMG-CoA synthase) (3-hydroxy-3-methylglutaryl coenzyme A synthase). | |||||
|
KAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.020206 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.001222 (rank : 152) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
OSBL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.010242 (rank : 135) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.015045 (rank : 126) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.018197 (rank : 124) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
PMS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.020343 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
SHD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.022387 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.028987 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.026428 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.010394 (rank : 133) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
HSN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.015001 (rank : 127) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IFS6 | Gene names | Hsn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.009934 (rank : 137) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.007348 (rank : 140) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WDR22_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.005963 (rank : 143) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |
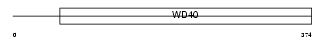
|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.010382 (rank : 134) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | -0.000940 (rank : 155) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.013880 (rank : 129) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.007174 (rank : 141) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.005684 (rank : 144) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | -0.000490 (rank : 153) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.004926 (rank : 145) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.008163 (rank : 139) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.010037 (rank : 136) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.017021 (rank : 125) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
IKBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.006415 (rank : 142) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |
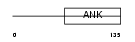
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
K1718_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.003540 (rank : 148) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.004637 (rank : 147) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.018264 (rank : 123) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.001465 (rank : 151) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.011888 (rank : 131) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.010413 (rank : 132) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
USH2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | -0.001234 (rank : 156) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
ATCAY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.060630 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WG3, Q8NAQ2, Q8TAQ3, Q96HC6, Q96JF5 | Gene names | ATCAY, KIAA1872 | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin (Ataxia Cayman type protein) (BNIP-H). | |||||
|
ATCAY_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.061413 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |
|
|||||
| Description | Caytaxin. | |||||
|
BNIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.064367 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12982 | Gene names | BNIP2, NIP2 | |||
|
Domain Architecture |
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
BNIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.064227 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54940 | Gene names | Bnip2, Nip2l | |||
|
Domain Architecture |
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
BNIPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.059199 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
BNIPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.059445 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JU7, Q923E3 | Gene names | Bnipl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
CENA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.051804 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.056380 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.069126 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.073815 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.068330 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CIP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.056815 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050656 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.055193 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.054490 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052179 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.074671 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
FA13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.075633 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.058898 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.057807 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
GRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.056050 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
Domain Architecture |
|
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.052767 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.053153 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62993, P29354, Q14450, Q63057, Q63059 | Gene names | GRB2, ASH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2) (Protein Ash). | |||||
|
GRB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.053145 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60631, Q61240 | Gene names | Grb2 | |||
|
Domain Architecture |
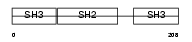
|
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2). | |||||
|
GRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.072947 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
NCF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050334 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97369 | Gene names | Ncf4 | |||
|
Domain Architecture |
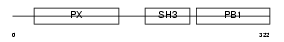
|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
NCK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.061328 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |
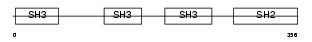
|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
P85B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.078580 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.052983 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PCTL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.082208 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |
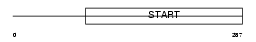
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
PCTL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.083138 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PDZD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.052734 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.054483 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.053100 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.052958 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.054714 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.054565 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051314 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050197 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
VAV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.051324 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.050802 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VINEX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.053232 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.994230 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.809581 (rank : 3) | θ value | 1.80466e-16 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.799530 (rank : 4) | θ value | 2.69047e-20 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.790769 (rank : 5) | θ value | 3.07829e-16 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.789867 (rank : 6) | θ value | 7.32683e-18 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.789746 (rank : 7) | θ value | 3.89403e-11 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.785582 (rank : 8) | θ value | 5.60996e-18 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.783693 (rank : 9) | θ value | 1.24977e-17 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.779891 (rank : 10) | θ value | 1.42992e-13 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.778129 (rank : 11) | θ value | 7.09661e-13 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.777791 (rank : 12) | θ value | 1.86753e-13 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.775499 (rank : 13) | θ value | 3.52202e-12 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.772893 (rank : 14) | θ value | 2.60593e-15 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.768998 (rank : 15) | θ value | 2.69671e-12 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.768869 (rank : 16) | θ value | 1.16975e-15 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.746671 (rank : 17) | θ value | 1.33837e-11 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.746619 (rank : 18) | θ value | 4.02038e-16 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.741170 (rank : 19) | θ value | 4.44505e-15 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.736290 (rank : 20) | θ value | 1.63225e-17 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.733934 (rank : 21) | θ value | 1.92812e-18 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.733744 (rank : 22) | θ value | 5.43371e-13 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.731686 (rank : 23) | θ value | 1.02475e-11 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.723863 (rank : 24) | θ value | 5.07402e-19 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.723168 (rank : 25) | θ value | 3.07829e-16 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.722793 (rank : 26) | θ value | 7.82807e-20 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.720618 (rank : 27) | θ value | 9.90251e-15 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.719414 (rank : 28) | θ value | 2.06002e-20 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.718867 (rank : 29) | θ value | 8.67504e-11 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.717573 (rank : 30) | θ value | 4.30538e-10 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.715677 (rank : 31) | θ value | 3.40345e-15 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
CHIO_HUMAN
|
||||||
| NC score | 0.715408 (rank : 32) | θ value | 5.25075e-16 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
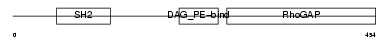
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.710916 (rank : 33) | θ value | 5.07402e-19 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.710661 (rank : 34) | θ value | 4.44505e-15 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.710540 (rank : 35) | θ value | 1.133e-10 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.701657 (rank : 36) | θ value | 3.07829e-16 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.699747 (rank : 37) | θ value | 4.02038e-16 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.697146 (rank : 38) | θ value | 2.13673e-09 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.693040 (rank : 39) | θ value | 2.20605e-14 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.692808 (rank : 40) | θ value | 2.60593e-15 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.692204 (rank : 41) | θ value | 1.86753e-13 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.691475 (rank : 42) | θ value | 3.40345e-15 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.690301 (rank : 43) | θ value | 9.26847e-13 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.688818 (rank : 44) | θ value | 3.18553e-13 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.688082 (rank : 45) | θ value | 5.43371e-13 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.683730 (rank : 46) | θ value | 3.76295e-14 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.659217 (rank : 47) | θ value | 9.90251e-15 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.645732 (rank : 48) | θ value | 4.16044e-13 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.643785 (rank : 49) | θ value | 2.43908e-13 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.624449 (rank : 50) | θ value | 1.33837e-11 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.598379 (rank : 51) | θ value | 4.16044e-13 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.587288 (rank : 52) | θ value | 1.58096e-12 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.555386 (rank : 53) | θ value | 8.40245e-06 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.543386 (rank : 54) | θ value | 8.40245e-06 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.532979 (rank : 55) | θ value | 6.64225e-11 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.514064 (rank : 56) | θ value | 6.64225e-11 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.413929 (rank : 57) | θ value | 8.95645e-16 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.408678 (rank : 58) | θ value | 6.85773e-16 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.350449 (rank : 59) | θ value | 0.000602161 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
OCRL_HUMAN
|
||||||
| NC score | 0.344616 (rank : 60) | θ value | 8.40245e-06 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.336942 (rank : 61) | θ value | 0.0113563 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.335745 (rank : 62) | θ value | 0.125558 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
PCTL_MOUSE
|
||||||
| NC score | 0.083138 (rank : 63) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PCTL_HUMAN
|
||||||
| NC score | 0.082208 (rank : 64) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.078580 (rank : 65) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
FA13A_MOUSE
|
||||||
| NC score | 0.075633 (rank : 66) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.074671 (rank : 67) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.073815 (rank : 68) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
DYDC2_HUMAN
|
||||||
| NC score | 0.073722 (rank : 69) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
GRP3_HUMAN
|
||||||
| NC score | 0.072947 (rank : 70) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.069126 (rank : 71) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.068330 (rank : 72) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
BNIP2_HUMAN
|
||||||
| NC score | 0.064367 (rank : 73) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12982 | Gene names | BNIP2, NIP2 | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
BNIP2_MOUSE
|
||||||
| NC score | 0.064227 (rank : 74) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54940 | Gene names | Bnip2, Nip2l | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
ATCAY_MOUSE
|
||||||
| NC score | 0.061413 (rank : 75) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin. | |||||
|
NCK2_HUMAN
|
||||||
| NC score | 0.061328 (rank : 76) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
ATCAY_HUMAN
|
||||||
| NC score | 0.060630 (rank : 77) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WG3, Q8NAQ2, Q8TAQ3, Q96HC6, Q96JF5 | Gene names | ATCAY, KIAA1872 | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin (Ataxia Cayman type protein) (BNIP-H). | |||||
|
BNIPL_MOUSE
|
||||||
| NC score | 0.059445 (rank : 78) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JU7, Q923E3 | Gene names | Bnipl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
BNIPL_HUMAN
|
||||||
| NC score | 0.059199 (rank : 79) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.058898 (rank : 80) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.057807 (rank : 81) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
CIP4_HUMAN
|
||||||
| NC score | 0.056815 (rank : 82) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.056380 (rank : 83) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
GRAP_HUMAN
|
||||||
| NC score | 0.056050 (rank : 84) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adapter protein. | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.055497 (rank : 85) | θ value | 0.0330416 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.055193 (rank : 86) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.054714 (rank : 87) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.054565 (rank : 88) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.054490 (rank : 89) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.054483 (rank : 90) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
VINEX_MOUSE
|
||||||
| NC score | 0.053232 (rank : 91) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
GRB2_HUMAN
|
||||||
| NC score | 0.053153 (rank : 92) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62993, P29354, Q14450, Q63057, Q63059 | Gene names | GRB2, ASH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2) (Protein Ash). | |||||
|
GRB2_MOUSE
|
||||||
| NC score | 0.053145 (rank : 93) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60631, Q61240 | Gene names | Grb2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.053100 (rank : 94) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.052983 (rank : 95) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.052958 (rank : 96) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
GRAP_MOUSE
|
||||||
| NC score | 0.052767 (rank : 97) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
PDZD8_HUMAN
|
||||||
| NC score | 0.052734 (rank : 98) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.052179 (rank : 99) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.051804 (rank : 100) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.051324 (rank : 101) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.051314 (rank : 102) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.050802 (rank : 103) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.050656 (rank : 104) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
NCF4_MOUSE
|
||||||
| NC score | 0.050334 (rank : 105) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97369 | Gene names | Ncf4 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.050197 (rank : 106) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.046623 (rank : 107) | θ value | 0.0148317 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
DNMBP_MOUSE
|
||||||
| NC score | 0.044878 (rank : 108) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.042726 (rank : 109) | θ value | 0.21417 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.040775 (rank : 110) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
K1219_HUMAN
|
||||||
| NC score | 0.030614 (rank : 111) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.028987 (rank : 112) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.028548 (rank : 113) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.026699 (rank : 114) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.026428 (rank : 115) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.023217 (rank : 116) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.022591 (rank : 117) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SHD_HUMAN
|
||||||
| NC score | 0.022387 (rank : 118) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.021182 (rank : 119) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.020343 (rank : 120) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.020206 (rank : 121) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.018518 (rank : 122) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.018264 (rank : 123) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.018197 (rank : 124) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.017021 (rank : 125) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.015045 (rank : 126) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
HSN2_MOUSE
|
||||||
| NC score | 0.015001 (rank : 127) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IFS6 | Gene names | Hsn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein HSN2 precursor. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.014321 (rank : 128) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.013880 (rank : 129) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
HMCS1_MOUSE
|
||||||
| NC score | 0.012118 (rank : 130) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8JZK9, Q3UXI4 | Gene names | Hmgcs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hydroxymethylglutaryl-CoA synthase, cytoplasmic (EC 2.3.3.10) (HMG-CoA synthase) (3-hydroxy-3-methylglutaryl coenzyme A synthase). | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.011888 (rank : 131) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.010413 (rank : 132) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.010394 (rank : 133) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.010382 (rank : 134) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
OSBL3_MOUSE
|
||||||
| NC score | 0.010242 (rank : 135) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.010037 (rank : 136) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.009934 (rank : 137) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.009853 (rank : 138) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.008163 (rank : 139) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.007348 (rank : 140) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.007174 (rank : 141) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
IKBL_HUMAN
|
||||||
| NC score | 0.006415 (rank : 142) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
WDR22_HUMAN
|
||||||
| NC score | 0.005963 (rank : 143) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.005684 (rank : 144) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.004926 (rank : 145) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.004823 (rank : 146) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.004637 (rank : 147) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.003540 (rank : 148) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.003165 (rank : 149) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.002109 (rank : 150) | θ value | 0.21417 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.001465 (rank : 151) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.001222 (rank : 152) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | -0.000490 (rank : 153) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
ZSCA1_HUMAN
|
||||||
| NC score | -0.000613 (rank : 154) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBB4, Q6WLH8, Q86WS8 | Gene names | ZSCAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 1. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.000940 (rank : 155) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
USH2A_MOUSE
|
||||||
| NC score | -0.001234 (rank : 156) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||