Please be patient as the page loads
|
OCRL_HUMAN
|
||||||
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
I5P2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975892 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
SYNJ2_HUMAN
|
||||||
| θ value | 4.24587e-58 (rank : 3) | NC score | 0.766504 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
SYNJ2_MOUSE
|
||||||
| θ value | 8.00737e-57 (rank : 4) | NC score | 0.773088 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 4.85792e-54 (rank : 5) | NC score | 0.702311 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 5.37103e-53 (rank : 6) | NC score | 0.703498 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 5.95217e-44 (rank : 7) | NC score | 0.664747 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 5.95217e-44 (rank : 8) | NC score | 0.735614 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SKIP_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 9) | NC score | 0.797671 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C5L6, O09040 | Gene names | Skip, Pps | |||
|
Domain Architecture |
|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
SKIP_HUMAN
|
||||||
| θ value | 2.05049e-36 (rank : 10) | NC score | 0.796797 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BT40, Q15733, Q9NPJ5, Q9P2R5 | Gene names | SKIP, PPS | |||
|
Domain Architecture |

|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
INP5E_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 11) | NC score | 0.734694 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 9.54697e-26 (rank : 12) | NC score | 0.711803 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 13) | NC score | 0.384201 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 14) | NC score | 0.381499 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 15) | NC score | 0.364365 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CHIO_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 16) | NC score | 0.332259 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 17) | NC score | 0.346680 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CHIO_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 18) | NC score | 0.344269 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 19) | NC score | 0.321228 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 20) | NC score | 0.362415 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 21) | NC score | 0.360686 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 22) | NC score | 0.344616 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 23) | NC score | 0.317749 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 24) | NC score | 0.323130 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 25) | NC score | 0.340490 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 26) | NC score | 0.352211 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 27) | NC score | 0.360911 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 28) | NC score | 0.322177 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 29) | NC score | 0.354510 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 30) | NC score | 0.177514 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
I5P1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 31) | NC score | 0.246273 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14642, Q14640 | Gene names | INPP5A | |||
|
Domain Architecture |

|
|||||
| Description | Type I inositol-1,4,5-trisphosphate 5-phosphatase (EC 3.1.3.56) (5PTase). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.201868 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 33) | NC score | 0.323485 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 34) | NC score | 0.300028 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 35) | NC score | 0.331420 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 36) | NC score | 0.304116 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 37) | NC score | 0.301858 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
P85A_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 38) | NC score | 0.199835 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 39) | NC score | 0.306736 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 40) | NC score | 0.314315 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 41) | NC score | 0.304501 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 42) | NC score | 0.311430 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 43) | NC score | 0.169966 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 44) | NC score | 0.284636 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
CE005_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 45) | NC score | 0.281637 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 46) | NC score | 0.286167 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 47) | NC score | 0.306221 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 48) | NC score | 0.280779 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 49) | NC score | 0.313967 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 50) | NC score | 0.281142 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 51) | NC score | 0.283732 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG18_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 52) | NC score | 0.284740 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG07_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 53) | NC score | 0.234698 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 54) | NC score | 0.322843 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.212646 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.235138 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
PROL4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.076271 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16378, Q8NFB3 | Gene names | PRR4, LPRP, PROL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 4 precursor (Lacrimal proline-rich protein) (Nasopharyngeal carcinoma-associated proline-rich protein 4). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.283133 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.031958 (rank : 86) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.273566 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.056734 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.286606 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.268461 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.159616 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.166663 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
I27RA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.013485 (rank : 89) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWB1, O60624 | Gene names | IL27RA, CRL1, TCCR, WSX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-27 receptor alpha chain precursor (IL-27R-alpha) (WSX-1) (Type I T-cell cytokine receptor) (TCCR) (Protein CRL1). | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | -0.002308 (rank : 98) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.001669 (rank : 97) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
HXB4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.003925 (rank : 95) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17483, Q9NTA0 | Gene names | HOXB4, HOX2F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2F) (Hox-2.6). | |||||
|
HXB4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.004311 (rank : 94) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.015466 (rank : 88) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
ANR25_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.003858 (rank : 96) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.011702 (rank : 90) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.009551 (rank : 91) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.023988 (rank : 87) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.009318 (rank : 92) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
SIRT6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.008058 (rank : 93) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6T7, O75291, Q6IAF5, Q6PK99, Q8NCD2, Q9BSI5, Q9BWP3, Q9NRC7, Q9UQD1 | Gene names | SIRT6, SIR2L6 | |||
|
Domain Architecture |
|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.171173 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.174855 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CF155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.066848 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.056276 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CO039_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055303 (rank : 85) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.055651 (rank : 84) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.059932 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.058816 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
P85B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.058579 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.062739 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.228904 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.234061 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.300652 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.272443 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.249449 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SAC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.209137 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |
|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SAC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.210571 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
STA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.189633 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.184881 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.200359 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.199576 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
OCRL_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.975892 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
SKIP_MOUSE
|
||||||
| NC score | 0.797671 (rank : 3) | θ value | 7.52953e-39 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C5L6, O09040 | Gene names | Skip, Pps | |||
|
Domain Architecture |
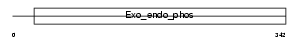
|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
SKIP_HUMAN
|
||||||
| NC score | 0.796797 (rank : 4) | θ value | 2.05049e-36 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BT40, Q15733, Q9NPJ5, Q9P2R5 | Gene names | SKIP, PPS | |||
|
Domain Architecture |

|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
SYNJ2_MOUSE
|
||||||
| NC score | 0.773088 (rank : 5) | θ value | 8.00737e-57 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
SYNJ2_HUMAN
|
||||||
| NC score | 0.766504 (rank : 6) | θ value | 4.24587e-58 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.735614 (rank : 7) | θ value | 5.95217e-44 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
INP5E_MOUSE
|
||||||
| NC score | 0.734694 (rank : 8) | θ value | 5.59698e-26 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.711803 (rank : 9) | θ value | 9.54697e-26 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.703498 (rank : 10) | θ value | 5.37103e-53 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.702311 (rank : 11) | θ value | 4.85792e-54 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.664747 (rank : 12) | θ value | 5.95217e-44 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.384201 (rank : 13) | θ value | 6.21693e-09 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.381499 (rank : 14) | θ value | 5.26297e-08 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.364365 (rank : 15) | θ value | 8.97725e-08 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.362415 (rank : 16) | θ value | 3.77169e-06 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.360911 (rank : 17) | θ value | 2.44474e-05 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.360686 (rank : 18) | θ value | 4.92598e-06 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.354510 (rank : 19) | θ value | 5.44631e-05 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.352211 (rank : 20) | θ value | 2.44474e-05 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.346680 (rank : 21) | θ value | 3.41135e-07 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.344616 (rank : 22) | θ value | 8.40245e-06 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.344269 (rank : 23) | θ value | 4.45536e-07 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.340490 (rank : 24) | θ value | 1.43324e-05 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
CHIO_HUMAN
|
||||||
| NC score | 0.332259 (rank : 25) | θ value | 1.53129e-07 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
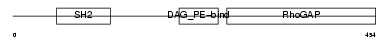
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.331420 (rank : 26) | θ value | 9.29e-05 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.323485 (rank : 27) | θ value | 9.29e-05 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.323130 (rank : 28) | θ value | 1.09739e-05 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.322843 (rank : 29) | θ value | 0.0563607 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.322177 (rank : 30) | θ value | 4.1701e-05 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.321228 (rank : 31) | θ value | 2.88788e-06 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.317749 (rank : 32) | θ value | 8.40245e-06 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.314315 (rank : 33) | θ value | 0.000461057 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.313967 (rank : 34) | θ value | 0.0148317 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.311430 (rank : 35) | θ value | 0.00102713 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.306736 (rank : 36) | θ value | 0.00020696 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.306221 (rank : 37) | θ value | 0.0113563 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.304501 (rank : 38) | θ value | 0.000786445 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.304116 (rank : 39) | θ value | 0.000121331 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.301858 (rank : 40) | θ value | 0.000158464 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.300652 (rank : 41) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.300028 (rank : 42) | θ value | 9.29e-05 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.286606 (rank : 43) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.286167 (rank : 44) | θ value | 0.0113563 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.284740 (rank : 45) | θ value | 0.0431538 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.284636 (rank : 46) | θ value | 0.00665767 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.283732 (rank : 47) | θ value | 0.0252991 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.283133 (rank : 48) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.281637 (rank : 49) | θ value | 0.00869519 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.281142 (rank : 50) | θ value | 0.0252991 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.280779 (rank : 51) | θ value | 0.0148317 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.273566 (rank : 52) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.272443 (rank : 53) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.268461 (rank : 54) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.249449 (rank : 55) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
I5P1_HUMAN
|
||||||
| NC score | 0.246273 (rank : 56) | θ value | 7.1131e-05 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14642, Q14640 | Gene names | INPP5A | |||
|
Domain Architecture |

|
|||||
| Description | Type I inositol-1,4,5-trisphosphate 5-phosphatase (EC 3.1.3.56) (5PTase). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.235138 (rank : 57) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.234698 (rank : 58) | θ value | 0.0563607 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.234061 (rank : 59) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.228904 (rank : 60) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.212646 (rank : 61) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
SAC3_MOUSE
|
||||||
| NC score | 0.210571 (rank : 62) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SAC3_HUMAN
|
||||||
| NC score | 0.209137 (rank : 63) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |

|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.201868 (rank : 64) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.200359 (rank : 65) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.199835 (rank : 66) | θ value | 0.00020696 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.199576 (rank : 67) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.189633 (rank : 68) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.184881 (rank : 69) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.177514 (rank : 70) | θ value | 5.44631e-05 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.174855 (rank : 71) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.171173 (rank : 72) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.169966 (rank : 73) | θ value | 0.00102713 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.166663 (rank : 74) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.159616 (rank : 75) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
PROL4_HUMAN
|
||||||
| NC score | 0.076271 (rank : 76) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16378, Q8NFB3 | Gene names | PRR4, LPRP, PROL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 4 precursor (Lacrimal proline-rich protein) (Nasopharyngeal carcinoma-associated proline-rich protein 4). | |||||
|
CF155_HUMAN
|
||||||
| NC score | 0.066848 (rank : 77) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.062739 (rank : 78) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.059932 (rank : 79) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.058816 (rank : 80) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.058579 (rank : 81) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.056734 (rank : 82) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.056276 (rank : 83) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.055651 (rank : 84) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.055303 (rank : 85) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.031958 (rank : 86) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.023988 (rank : 87) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.015466 (rank : 88) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
I27RA_HUMAN
|
||||||
| NC score | 0.013485 (rank : 89) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWB1, O60624 | Gene names | IL27RA, CRL1, TCCR, WSX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-27 receptor alpha chain precursor (IL-27R-alpha) (WSX-1) (Type I T-cell cytokine receptor) (TCCR) (Protein CRL1). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.011702 (rank : 90) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.009551 (rank : 91) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.009318 (rank : 92) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
SIRT6_HUMAN
|
||||||
| NC score | 0.008058 (rank : 93) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6T7, O75291, Q6IAF5, Q6PK99, Q8NCD2, Q9BSI5, Q9BWP3, Q9NRC7, Q9UQD1 | Gene names | SIRT6, SIR2L6 | |||
|
Domain Architecture |

|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
HXB4_MOUSE
|
||||||
| NC score | 0.004311 (rank : 94) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
HXB4_HUMAN
|
||||||
| NC score | 0.003925 (rank : 95) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17483, Q9NTA0 | Gene names | HOXB4, HOX2F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2F) (Hox-2.6). | |||||
|
ANR25_HUMAN
|
||||||
| NC score | 0.003858 (rank : 96) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.001669 (rank : 97) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | -0.002308 (rank : 98) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||