Please be patient as the page loads
|
STAR8_MOUSE
|
||||||
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RHG07_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.970772 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.971116 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.971763 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.971905 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.986481 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 7) | NC score | 0.704521 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 8) | NC score | 0.701657 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 9) | NC score | 0.344802 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 10) | NC score | 0.717371 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 11) | NC score | 0.349084 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 12) | NC score | 0.665856 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 13) | NC score | 0.637477 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 14) | NC score | 0.600281 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 15) | NC score | 0.615046 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 16) | NC score | 0.678548 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG18_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 17) | NC score | 0.706411 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 18) | NC score | 0.582397 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
BCR_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 19) | NC score | 0.568653 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 20) | NC score | 0.572559 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 21) | NC score | 0.592287 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 22) | NC score | 0.534587 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CE005_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 23) | NC score | 0.535686 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 24) | NC score | 0.631568 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 25) | NC score | 0.530580 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 26) | NC score | 0.598285 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 27) | NC score | 0.562711 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 28) | NC score | 0.555544 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 29) | NC score | 0.630708 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 30) | NC score | 0.489457 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 31) | NC score | 0.476982 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 32) | NC score | 0.625705 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 33) | NC score | 0.614521 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 34) | NC score | 0.627124 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 35) | NC score | 0.549651 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 36) | NC score | 0.622737 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 37) | NC score | 0.556607 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 38) | NC score | 0.464689 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 39) | NC score | 0.554388 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIO_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 40) | NC score | 0.579430 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
CHIO_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 41) | NC score | 0.549275 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
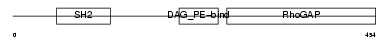
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 42) | NC score | 0.550209 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 43) | NC score | 0.621074 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 44) | NC score | 0.573282 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 45) | NC score | 0.567850 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 46) | NC score | 0.545703 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 47) | NC score | 0.584488 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 48) | NC score | 0.588802 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 49) | NC score | 0.578724 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 50) | NC score | 0.569354 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 51) | NC score | 0.533125 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 52) | NC score | 0.556635 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 53) | NC score | 0.577495 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
PCTL_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 54) | NC score | 0.317585 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |
|
|||||
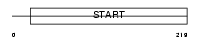
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PCTL_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 55) | NC score | 0.314960 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |
|
|||||
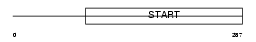
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 56) | NC score | 0.564238 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
STAR7_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 57) | NC score | 0.192971 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |
|
|||||
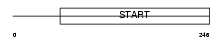
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
STAR7_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 58) | NC score | 0.175519 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |
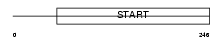
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 59) | NC score | 0.253406 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 60) | NC score | 0.255984 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
CAD24_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.006458 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86UP0, Q86UP1, Q9NT84 | Gene names | CDH24, CDH11L | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-24 precursor. | |||||
|
K1688_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 62) | NC score | 0.402713 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| θ value | 0.125558 (rank : 63) | NC score | 0.391322 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.125558 (rank : 64) | NC score | 0.024138 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
I5P2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.235827 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
EDAD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.031386 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHX2 | Gene names | Edaradd, Cr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectodysplasin A receptor-associated adapter protein (EDAR-associated death domain protein) (Crinkled protein). | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.008938 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
SALL4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.000238 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJQ4 | Gene names | SALL4 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 4 (Zinc finger protein SALL4). | |||||
|
GAGC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.025637 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60829, Q6IBI1 | Gene names | PAGE4, GAGEC1 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen family C member 1 (Prostate-associated gene 4 protein) (PAGE-4) (PAGE-1) (GAGE-9). | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | -0.000550 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ACO11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.048841 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXI4, O75187, Q96DI1, Q9H883 | Gene names | ACOT11, BFIT, KIAA0707, THEA | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
ACO12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.050313 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WYK0 | Gene names | ACOT12, CACH, CACH1, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (hCACH-1) (START domain-containing protein 12) (StARD12). | |||||
|
CENPC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.019291 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
CLK4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.001075 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAZ1 | Gene names | CLK4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK4 (EC 2.7.12.1) (CDC-like kinase 4). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.006434 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ACO12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.066874 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBK0, Q544M5, Q8R108 | Gene names | Acot12, Cach, Cach1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (mCACH-1). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.005368 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.006449 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
HIPK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.000496 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZR5, O88905, Q99P45, Q99P46, Q9D2E6, Q9D474, Q9EQL2, Q9QZR4 | Gene names | Hipk2, Nbak1, Stank | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (Nuclear body- associated kinase 1) (Sialophorin tail-associated nuclear serine/threonine-protein kinase). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.015613 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.009366 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
MDR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.002545 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21447 | Gene names | Abcb1a, Abcb4, Mdr1a, Mdr3, Pgy-3, Pgy3 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 3 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1A) (P-glycoprotein 3) (MDR1A). | |||||
|
RFTN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.011903 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q52LD8, Q2TA69, Q53QE0, Q53SE1, Q96NM3 | Gene names | RFTN2, C2orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin-2 (Raft-linking protein 2). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.013770 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
TBX18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.004031 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
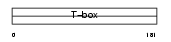
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
ALKB7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.014625 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BT30, Q53FF3 | Gene names | ALKBH7, SPATA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkylated repair protein alkB homolog 7 precursor (Spermatogenesis- associated protein 11) (Spermatogenesis cell proliferation-related protein). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.003879 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MANA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.012281 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924M7 | Gene names | Mpi, Mpi1, Pmi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannose-6-phosphate isomerase (EC 5.3.1.8) (Phosphomannose isomerase) (PMI) (Phosphohexomutase). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.015638 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.002558 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.003645 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
HIPK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.000072 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H2X6, Q75MR7, Q8WWI4, Q9H2Y1 | Gene names | HIPK2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (hHIPk2). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.005184 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.009238 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.007264 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.008084 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TSH3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.006385 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | -0.002115 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CLK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | -0.000205 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35493, O35721, Q8CEU9, Q99LU6 | Gene names | Clk4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK4 (EC 2.7.12.1) (CDC-like kinase 4). | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.008427 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.016436 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.320435 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.336069 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.060298 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
FA13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.061151 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
OCRL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.199576 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
P85B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.059153 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.986481 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.971905 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.971763 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.971116 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.970772 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.717371 (rank : 7) | θ value | 1.68911e-14 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.706411 (rank : 8) | θ value | 3.52202e-12 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.704521 (rank : 9) | θ value | 1.80466e-16 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.701657 (rank : 10) | θ value | 3.07829e-16 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.678548 (rank : 11) | θ value | 3.52202e-12 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.665856 (rank : 12) | θ value | 2.88119e-14 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.637477 (rank : 13) | θ value | 2.88119e-14 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.631568 (rank : 14) | θ value | 1.9326e-10 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.630708 (rank : 15) | θ value | 5.62301e-10 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.627124 (rank : 16) | θ value | 1.63604e-09 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.625705 (rank : 17) | θ value | 9.59137e-10 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.622737 (rank : 18) | θ value | 2.79066e-09 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.621074 (rank : 19) | θ value | 3.41135e-07 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.615046 (rank : 20) | θ value | 5.43371e-13 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.614521 (rank : 21) | θ value | 1.63604e-09 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.600281 (rank : 22) | θ value | 6.41864e-14 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.598285 (rank : 23) | θ value | 1.9326e-10 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.592287 (rank : 24) | θ value | 3.89403e-11 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.588802 (rank : 25) | θ value | 1.29631e-06 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.584488 (rank : 26) | θ value | 1.29631e-06 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.582397 (rank : 27) | θ value | 4.59992e-12 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.579430 (rank : 28) | θ value | 1.80886e-08 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.578724 (rank : 29) | θ value | 1.29631e-06 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.577495 (rank : 30) | θ value | 8.40245e-06 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.573282 (rank : 31) | θ value | 5.81887e-07 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.572559 (rank : 32) | θ value | 2.28291e-11 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.569354 (rank : 33) | θ value | 2.21117e-06 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.568653 (rank : 34) | θ value | 6.00763e-12 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.567850 (rank : 35) | θ value | 9.92553e-07 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.564238 (rank : 36) | θ value | 7.1131e-05 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.562711 (rank : 37) | θ value | 2.52405e-10 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.556635 (rank : 38) | θ value | 8.40245e-06 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.556607 (rank : 39) | θ value | 2.79066e-09 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.555544 (rank : 40) | θ value | 5.62301e-10 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.554388 (rank : 41) | θ value | 1.80886e-08 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.550209 (rank : 42) | θ value | 6.87365e-08 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.549651 (rank : 43) | θ value | 1.63604e-09 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
CHIO_HUMAN
|
||||||
| NC score | 0.549275 (rank : 44) | θ value | 5.26297e-08 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
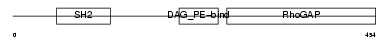
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.545703 (rank : 45) | θ value | 9.92553e-07 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.535686 (rank : 46) | θ value | 1.47974e-10 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.534587 (rank : 47) | θ value | 8.67504e-11 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.533125 (rank : 48) | θ value | 4.92598e-06 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.530580 (rank : 49) | θ value | 1.9326e-10 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.489457 (rank : 50) | θ value | 7.34386e-10 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.476982 (rank : 51) | θ value | 9.59137e-10 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.464689 (rank : 52) | θ value | 3.64472e-09 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.402713 (rank : 53) | θ value | 0.0961366 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.391322 (rank : 54) | θ value | 0.125558 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.349084 (rank : 55) | θ value | 2.20605e-14 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.344802 (rank : 56) | θ value | 4.44505e-15 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.336069 (rank : 57) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.320435 (rank : 58) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
PCTL_MOUSE
|
||||||
| NC score | 0.317585 (rank : 59) | θ value | 3.19293e-05 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PCTL_HUMAN
|
||||||
| NC score | 0.314960 (rank : 60) | θ value | 4.1701e-05 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.255984 (rank : 61) | θ value | 0.0330416 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.253406 (rank : 62) | θ value | 0.0113563 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.235827 (rank : 63) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| NC score | 0.199576 (rank : 64) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
STAR7_HUMAN
|
||||||
| NC score | 0.192971 (rank : 65) | θ value | 0.00102713 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
STAR7_MOUSE
|
||||||
| NC score | 0.175519 (rank : 66) | θ value | 0.00390308 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
ACO12_MOUSE
|
||||||
| NC score | 0.066874 (rank : 67) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBK0, Q544M5, Q8R108 | Gene names | Acot12, Cach, Cach1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (mCACH-1). | |||||
|
FA13A_MOUSE
|
||||||
| NC score | 0.061151 (rank : 68) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.060298 (rank : 69) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.059153 (rank : 70) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
ACO12_HUMAN
|
||||||
| NC score | 0.050313 (rank : 71) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WYK0 | Gene names | ACOT12, CACH, CACH1, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (hCACH-1) (START domain-containing protein 12) (StARD12). | |||||
|
ACO11_HUMAN
|
||||||
| NC score | 0.048841 (rank : 72) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXI4, O75187, Q96DI1, Q9H883 | Gene names | ACOT11, BFIT, KIAA0707, THEA | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
EDAD_MOUSE
|
||||||
| NC score | 0.031386 (rank : 73) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHX2 | Gene names | Edaradd, Cr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectodysplasin A receptor-associated adapter protein (EDAR-associated death domain protein) (Crinkled protein). | |||||
|
GAGC1_HUMAN
|
||||||
| NC score | 0.025637 (rank : 74) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60829, Q6IBI1 | Gene names | PAGE4, GAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen family C member 1 (Prostate-associated gene 4 protein) (PAGE-4) (PAGE-1) (GAGE-9). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.024138 (rank : 75) | θ value | 0.125558 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CENPC_MOUSE
|
||||||
| NC score | 0.019291 (rank : 76) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.016436 (rank : 77) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.015638 (rank : 78) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.015613 (rank : 79) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ALKB7_HUMAN
|
||||||
| NC score | 0.014625 (rank : 80) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BT30, Q53FF3 | Gene names | ALKBH7, SPATA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkylated repair protein alkB homolog 7 precursor (Spermatogenesis- associated protein 11) (Spermatogenesis cell proliferation-related protein). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.013770 (rank : 81) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
MANA_MOUSE
|
||||||
| NC score | 0.012281 (rank : 82) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924M7 | Gene names | Mpi, Mpi1, Pmi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannose-6-phosphate isomerase (EC 5.3.1.8) (Phosphomannose isomerase) (PMI) (Phosphohexomutase). | |||||
|
RFTN2_HUMAN
|
||||||
| NC score | 0.011903 (rank : 83) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q52LD8, Q2TA69, Q53QE0, Q53SE1, Q96NM3 | Gene names | RFTN2, C2orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin-2 (Raft-linking protein 2). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.009366 (rank : 84) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.009238 (rank : 85) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.008938 (rank : 86) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.008427 (rank : 87) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.008084 (rank : 88) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.007264 (rank : 89) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
CAD24_HUMAN
|
||||||
| NC score | 0.006458 (rank : 90) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86UP0, Q86UP1, Q9NT84 | Gene names | CDH24, CDH11L | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-24 precursor. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.006449 (rank : 91) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.006434 (rank : 92) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
TSH3_HUMAN
|
||||||
| NC score | 0.006385 (rank : 93) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.005368 (rank : 94) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.005184 (rank : 95) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.004031 (rank : 96) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.003879 (rank : 97) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.003645 (rank : 98) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.002558 (rank : 99) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MDR3_MOUSE
|
||||||
| NC score | 0.002545 (rank : 100) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21447 | Gene names | Abcb1a, Abcb4, Mdr1a, Mdr3, Pgy-3, Pgy3 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 3 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1A) (P-glycoprotein 3) (MDR1A). | |||||
|
CLK4_HUMAN
|
||||||
| NC score | 0.001075 (rank : 101) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAZ1 | Gene names | CLK4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK4 (EC 2.7.12.1) (CDC-like kinase 4). | |||||
|
HIPK2_MOUSE
|
||||||
| NC score | 0.000496 (rank : 102) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZR5, O88905, Q99P45, Q99P46, Q9D2E6, Q9D474, Q9EQL2, Q9QZR4 | Gene names | Hipk2, Nbak1, Stank | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (Nuclear body- associated kinase 1) (Sialophorin tail-associated nuclear serine/threonine-protein kinase). | |||||
|
SALL4_HUMAN
|
||||||
| NC score | 0.000238 (rank : 103) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJQ4 | Gene names | SALL4 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 4 (Zinc finger protein SALL4). | |||||
|
HIPK2_HUMAN
|
||||||
| NC score | 0.000072 (rank : 104) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H2X6, Q75MR7, Q8WWI4, Q9H2Y1 | Gene names | HIPK2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (hHIPk2). | |||||
|
CLK4_MOUSE
|
||||||
| NC score | -0.000205 (rank : 105) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35493, O35721, Q8CEU9, Q99LU6 | Gene names | Clk4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK4 (EC 2.7.12.1) (CDC-like kinase 4). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | -0.000550 (rank : 106) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | -0.002115 (rank : 107) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||