Please be patient as the page loads
|
FA13A_MOUSE
|
||||||
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FA13A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.981079 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
FA13A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 3.09092e-117 (rank : 3) | NC score | 0.698053 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 3.53647e-113 (rank : 4) | NC score | 0.695623 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
FA13C_HUMAN
|
||||||
| θ value | 3.974e-56 (rank : 5) | NC score | 0.843498 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_MOUSE
|
||||||
| θ value | 2.66562e-52 (rank : 6) | NC score | 0.845183 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 7) | NC score | 0.033487 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.073921 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
LAMA4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 9) | NC score | 0.044189 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.057384 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.044131 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.032877 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.021618 (rank : 98) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.059749 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PREP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.067377 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.053056 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.056025 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.048202 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TIM44_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.050633 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
K1C17_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.032518 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04695 | Gene names | KRT17 | |||
|
Domain Architecture |
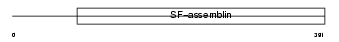
|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17) (39.1). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.065851 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.058800 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.050731 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
APOA4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.066644 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |
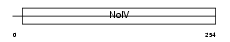
|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.049919 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.077422 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.020489 (rank : 99) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.048966 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
K1C13_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.028808 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |
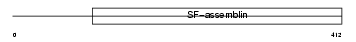
|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
K1C23_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.030456 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99PS0 | Gene names | Krt23, Haik1, Krt1-23 | |||
|
Domain Architecture |
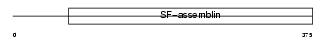
|
|||||
| Description | Keratin, type I cytoskeletal 23 (Cytokeratin-23) (CK-23) (Keratin-23) (K23). | |||||
|
KU70_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.036799 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23475, O88212, Q62027, Q62382, Q62453 | Gene names | Xrcc6, G22p1, Ku70 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Ku autoantigen protein p70 homolog) (Ku70) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA- repair protein XRCC6). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.048617 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.024021 (rank : 97) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.039359 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.046188 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.053551 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.049367 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.050018 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.020213 (rank : 100) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
K1C17_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.030147 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||
|
LRRC7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.007434 (rank : 118) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
MCM5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.017643 (rank : 105) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.044171 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.015248 (rank : 109) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.045826 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
IL16_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.015766 (rank : 108) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.029732 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
NOC3L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.026658 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.019988 (rank : 101) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZC11A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.026693 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.029014 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.034888 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.005507 (rank : 120) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.034521 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.018837 (rank : 103) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.044830 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.019602 (rank : 102) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
KINH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.037287 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.038947 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.027831 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.013445 (rank : 111) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.032875 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCD70_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.017060 (rank : 106) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9B0, Q9D9H6 | Gene names | Ccdc70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 70 precursor. | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.009346 (rank : 116) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.009341 (rank : 117) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.045647 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
K0460_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.016638 (rank : 107) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.031950 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MCM5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.014557 (rank : 110) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MUTED_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.032261 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDH9, Q5THS1, Q68D56, Q8N5F9, Q9NU16 | Gene names | MUTED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muted protein homolog. | |||||
|
NARGL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.013197 (rank : 114) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
PI52B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.013333 (rank : 112) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78356, Q5U0E8, Q8TBP2 | Gene names | PIP5K2B | |||
|
Domain Architecture |
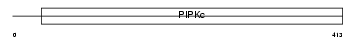
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
PI52B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.013214 (rank : 113) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XI4, Q8VHB2 | Gene names | Pip5k2b | |||
|
Domain Architecture |
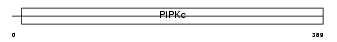
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.034817 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.027728 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.042585 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.040238 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INOC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005769 (rank : 119) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
K1C19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.024919 (rank : 95) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19001 | Gene names | Krt19, Krt1-19 | |||
|
Domain Architecture |
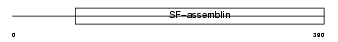
|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.017883 (rank : 104) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
XPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.013018 (rank : 115) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01831, Q96AX0 | Gene names | XPC, XPCC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells (Xeroderma pigmentosum group C-complementing protein) (p125). | |||||
|
ZBT26_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | -0.001688 (rank : 121) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCK0, Q8WTR1 | Gene names | ZBTB26, KIAA1572, ZNF481 | |||
|
Domain Architecture |
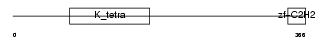
|
|||||
| Description | Zinc finger and BTB domain-containing protein 26 (Zinc finger protein 481) (Zinc finger protein Bioref). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.024094 (rank : 96) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ABR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.054039 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
CEND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.053527 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CHIO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.052624 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051548 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.076333 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.076337 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.065370 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
K1688_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.063917 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.066954 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.059808 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.058055 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.074735 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.071703 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.079979 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.062929 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.090347 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.069834 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.068880 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.075633 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.075376 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG07_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.054327 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058937 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.068533 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.071856 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG09_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.058765 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053795 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.053703 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.066038 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.078884 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.093015 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.092507 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.055997 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.053103 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050382 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.059912 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.061151 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.055775 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.063139 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
FA13A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.981079 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.845183 (rank : 3) | θ value | 2.66562e-52 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.843498 (rank : 4) | θ value | 3.974e-56 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.698053 (rank : 5) | θ value | 3.09092e-117 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.695623 (rank : 6) | θ value | 3.53647e-113 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.093015 (rank : 7) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.092507 (rank : 8) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.090347 (rank : 9) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.079979 (rank : 10) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.078884 (rank : 11) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.077422 (rank : 12) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.076337 (rank : 13) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.076333 (rank : 14) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.075633 (rank : 15) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.075376 (rank : 16) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.074735 (rank : 17) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.073921 (rank : 18) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.071856 (rank : 19) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.071703 (rank : 20) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.069834 (rank : 21) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.068880 (rank : 22) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.068533 (rank : 23) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
PREP_HUMAN
|
||||||
| NC score | 0.067377 (rank : 24) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.066954 (rank : 25) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
APOA4_MOUSE
|
||||||
| NC score | 0.066644 (rank : 26) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.066038 (rank : 27) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.065851 (rank : 28) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.065370 (rank : 29) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.063917 (rank : 30) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.063139 (rank : 31) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.062929 (rank : 32) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.061151 (rank : 33) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.059912 (rank : 34) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.059808 (rank : 35) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.059749 (rank : 36) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.058937 (rank : 37) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.058800 (rank : 38) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.058765 (rank : 39) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.058055 (rank : 40) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.057384 (rank : 41) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.056025 (rank : 42) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.055997 (rank : 43) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.055775 (rank : 44) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.054327 (rank : 45) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.054039 (rank : 46) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.053795 (rank : 47) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.053703 (rank : 48) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.053551 (rank : 49) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.053527 (rank : 50) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.053103 (rank : 51) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.053056 (rank : 52) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.052624 (rank : 53) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.051548 (rank : 54) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.050731 (rank : 55) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
TIM44_MOUSE
|
||||||
| NC score | 0.050633 (rank : 56) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.050382 (rank : 57) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.050018 (rank : 58) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.049919 (rank : 59) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.049367 (rank : 60) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.048966 (rank : 61) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.048617 (rank : 62) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.048202 (rank : 63) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.046188 (rank : 64) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.045826 (rank : 65) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.045647 (rank : 66) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.044830 (rank : 67) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
LAMA4_HUMAN
|
||||||
| NC score | 0.044189 (rank : 68) | θ value | 0.0148317 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.044171 (rank : 69) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.044131 (rank : 70) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.042585 (rank : 71) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.040238 (rank : 72) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.039359 (rank : 73) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.038947 (rank : 74) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.037287 (rank : 75) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KU70_MOUSE
|
||||||
| NC score | 0.036799 (rank : 76) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23475, O88212, Q62027, Q62382, Q62453 | Gene names | Xrcc6, G22p1, Ku70 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Ku autoantigen protein p70 homolog) (Ku70) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA- repair protein XRCC6). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.034888 (rank : 77) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.034817 (rank : 78) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.034521 (rank : 79) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.033487 (rank : 80) | θ value | 0.00175202 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.032877 (rank : 81) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.032875 (rank : 82) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
K1C17_HUMAN
|
||||||
| NC score | 0.032518 (rank : 83) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04695 | Gene names | KRT17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17) (39.1). | |||||
|
MUTED_HUMAN
|
||||||
| NC score | 0.032261 (rank : 84) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDH9, Q5THS1, Q68D56, Q8N5F9, Q9NU16 | Gene names | MUTED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muted protein homolog. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.031950 (rank : 85) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
K1C23_MOUSE
|
||||||
| NC score | 0.030456 (rank : 86) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99PS0 | Gene names | Krt23, Haik1, Krt1-23 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 23 (Cytokeratin-23) (CK-23) (Keratin-23) (K23). | |||||
|
K1C17_MOUSE
|
||||||
| NC score | 0.030147 (rank : 87) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.029732 (rank : 88) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.029014 (rank : 89) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
K1C13_HUMAN
|
||||||
| NC score | 0.028808 (rank : 90) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.027831 (rank : 91) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.027728 (rank : 92) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
ZC11A_HUMAN
|
||||||
| NC score | 0.026693 (rank : 93) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
NOC3L_HUMAN
|
||||||
| NC score | 0.026658 (rank : 94) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
K1C19_MOUSE
|
||||||
| NC score | 0.024919 (rank : 95) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19001 | Gene names | Krt19, Krt1-19 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.024094 (rank : 96) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.024021 (rank : 97) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.021618 (rank : 98) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.020489 (rank : 99) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.020213 (rank : 100) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.019988 (rank : 101) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.019602 (rank : 102) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.018837 (rank : 103) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.017883 (rank : 104) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
MCM5_HUMAN
|
||||||
| NC score | 0.017643 (rank : 105) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
CCD70_MOUSE
|
||||||
| NC score | 0.017060 (rank : 106) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9B0, Q9D9H6 | Gene names | Ccdc70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 70 precursor. | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.016638 (rank : 107) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
IL16_MOUSE
|
||||||
| NC score | 0.015766 (rank : 108) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.015248 (rank : 109) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
MCM5_MOUSE
|
||||||
| NC score | 0.014557 (rank : 110) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.013445 (rank : 111) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
PI52B_HUMAN
|
||||||
| NC score | 0.013333 (rank : 112) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78356, Q5U0E8, Q8TBP2 | Gene names | PIP5K2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
PI52B_MOUSE
|
||||||
| NC score | 0.013214 (rank : 113) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XI4, Q8VHB2 | Gene names | Pip5k2b | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
NARGL_MOUSE
|
||||||
| NC score | 0.013197 (rank : 114) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
XPC_HUMAN
|
||||||
| NC score | 0.013018 (rank : 115) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01831, Q96AX0 | Gene names | XPC, XPCC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells (Xeroderma pigmentosum group C-complementing protein) (p125). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.009346 (rank : 116) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.009341 (rank : 117) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
LRRC7_HUMAN
|
||||||
| NC score | 0.007434 (rank : 118) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.005769 (rank : 119) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.005507 (rank : 120) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ZBT26_HUMAN
|
||||||
| NC score | -0.001688 (rank : 121) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCK0, Q8WTR1 | Gene names | ZBTB26, KIAA1572, ZNF481 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 26 (Zinc finger protein 481) (Zinc finger protein Bioref). | |||||