Please be patient as the page loads
|
RHG18_MOUSE
|
||||||
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RHG18_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987326 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 3) | NC score | 0.768583 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 4) | NC score | 0.764212 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 5) | NC score | 0.725259 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 6) | NC score | 0.408863 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 7) | NC score | 0.413900 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 8) | NC score | 0.715514 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 9) | NC score | 0.722784 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 10) | NC score | 0.702675 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 11) | NC score | 0.761752 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 12) | NC score | 0.763882 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
STAR8_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 13) | NC score | 0.717371 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 14) | NC score | 0.692033 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 15) | NC score | 0.707968 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 16) | NC score | 0.754501 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 17) | NC score | 0.758644 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 18) | NC score | 0.760874 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 19) | NC score | 0.772393 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 20) | NC score | 0.771105 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 21) | NC score | 0.668006 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 22) | NC score | 0.775499 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 23) | NC score | 0.628032 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
RHG01_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 24) | NC score | 0.748252 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 25) | NC score | 0.774371 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 26) | NC score | 0.628752 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 27) | NC score | 0.728081 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 28) | NC score | 0.672450 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
CHIO_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 29) | NC score | 0.704471 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 30) | NC score | 0.728100 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 31) | NC score | 0.755113 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 32) | NC score | 0.663085 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 33) | NC score | 0.671732 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIO_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 34) | NC score | 0.671390 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
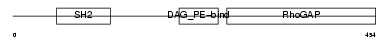
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 35) | NC score | 0.593873 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 36) | NC score | 0.717850 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 37) | NC score | 0.682380 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
K1688_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 38) | NC score | 0.580952 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 39) | NC score | 0.569126 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 40) | NC score | 0.671214 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 41) | NC score | 0.624974 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 42) | NC score | 0.703954 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 43) | NC score | 0.599229 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 44) | NC score | 0.740276 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 45) | NC score | 0.577395 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 46) | NC score | 0.686175 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 47) | NC score | 0.697045 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 48) | NC score | 0.696348 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 49) | NC score | 0.673764 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 50) | NC score | 0.499829 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 51) | NC score | 0.666313 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 52) | NC score | 0.479565 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 53) | NC score | 0.644116 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 54) | NC score | 0.626390 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 55) | NC score | 0.617356 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 56) | NC score | 0.608821 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 57) | NC score | 0.664448 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 58) | NC score | 0.600771 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 59) | NC score | 0.034917 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 60) | NC score | 0.034884 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
I5P2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 61) | NC score | 0.313786 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 62) | NC score | 0.284740 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
FACD2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 63) | NC score | 0.061017 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXW9, Q2LA86, Q69YP9, Q6PJN7, Q9BQ06, Q9H9T9 | Gene names | FANCD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein (Protein FACD2). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.301146 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
CENPQ_HUMAN
|
||||||
| θ value | 0.21417 (rank : 65) | NC score | 0.042372 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7L2Z9, Q6IN61, Q9NVS5 | Gene names | CENPQ, C6orf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 66) | NC score | 0.032216 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.028728 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 68) | NC score | 0.022359 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.011491 (rank : 122) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.036596 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.026465 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.036581 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.028621 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.008549 (rank : 127) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.003661 (rank : 131) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.015669 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.010850 (rank : 123) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WBP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.035709 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
XPO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.019977 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5F9 | Gene names | Xpo1, Crm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-1 (Exp1) (Chromosome region maintenance 1 protein homolog). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.016141 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.014132 (rank : 114) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.027218 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.053707 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
SLBP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.027323 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97440 | Gene names | Slbp, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.012556 (rank : 119) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.012003 (rank : 120) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
DREB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.016154 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
MCM4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.012613 (rank : 118) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.036897 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.007987 (rank : 129) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
RAE2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.014270 (rank : 113) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26374, Q9H1Y4 | Gene names | CHML, REP2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
DAPK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | -0.005536 (rank : 136) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54784 | Gene names | Dapk3, Zipk | |||
|
Domain Architecture |

|
|||||
| Description | Death-associated protein kinase 3 (EC 2.7.11.1) (DAP kinase 3) (DAP- like kinase) (Dlk) (ZIP-kinase). | |||||
|
NRK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | -0.004421 (rank : 135) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||
|
SLBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.024234 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14493 | Gene names | SLBP, HBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.015434 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
UT14B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.020822 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
ATS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.001171 (rank : 134) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
CCND3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.004546 (rank : 130) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |
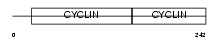
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CI039_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.015767 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.009678 (rank : 125) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
DREB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.013156 (rank : 117) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
GP73_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.013387 (rank : 116) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
KLHL6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.001261 (rank : 133) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6V595 | Gene names | Klhl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 6. | |||||
|
LAMB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.001820 (rank : 132) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.050813 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.013998 (rank : 115) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.008266 (rank : 128) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.011628 (rank : 121) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
VPS54_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.009032 (rank : 126) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SPW0, Q8BPB3, Q8CFZ7, Q8CHL5, Q8R3R4, Q8R3X1 | Gene names | Vps54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p homolog). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.010691 (rank : 124) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ATCAY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.052424 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WG3, Q8NAQ2, Q8TAQ3, Q96HC6, Q96JF5 | Gene names | ATCAY, KIAA1872 | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin (Ataxia Cayman type protein) (BNIP-H). | |||||
|
ATCAY_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052907 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |
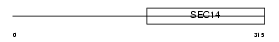
|
|||||
| Description | Caytaxin. | |||||
|
BNIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.055629 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12982 | Gene names | BNIP2, NIP2 | |||
|
Domain Architecture |
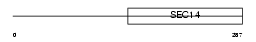
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
BNIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055547 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54940 | Gene names | Bnip2, Nip2l | |||
|
Domain Architecture |
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
BNIPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051209 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
BNIPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051399 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JU7, Q923E3 | Gene names | Bnipl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
CENA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052452 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.066336 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.072167 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.066946 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.051920 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.050017 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.078579 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
FA13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.078884 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.061349 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.059688 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
GRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.064409 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.054290 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
P85A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.313309 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.067284 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PCTL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.099299 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
PCTL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.100455 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051381 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.050266 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.051599 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.051817 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.987326 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.775499 (rank : 3) | θ value | 3.52202e-12 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.774371 (rank : 4) | θ value | 4.59992e-12 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.772393 (rank : 5) | θ value | 4.16044e-13 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.771105 (rank : 6) | θ value | 1.2105e-12 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.768583 (rank : 7) | θ value | 4.43474e-23 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.764212 (rank : 8) | θ value | 3.75424e-22 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.763882 (rank : 9) | θ value | 4.44505e-15 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.761752 (rank : 10) | θ value | 3.40345e-15 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.760874 (rank : 11) | θ value | 2.43908e-13 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.758644 (rank : 12) | θ value | 1.42992e-13 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.755113 (rank : 13) | θ value | 3.89403e-11 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.754501 (rank : 14) | θ value | 1.09485e-13 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.748252 (rank : 15) | θ value | 4.59992e-12 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.740276 (rank : 16) | θ value | 3.29651e-10 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.728100 (rank : 17) | θ value | 3.89403e-11 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.728081 (rank : 18) | θ value | 1.33837e-11 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.725259 (rank : 19) | θ value | 1.92812e-18 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.722784 (rank : 20) | θ value | 3.07829e-16 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.717850 (rank : 21) | θ value | 1.47974e-10 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.717371 (rank : 22) | θ value | 1.68911e-14 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.715514 (rank : 23) | θ value | 2.7842e-17 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.707968 (rank : 24) | θ value | 2.88119e-14 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.704471 (rank : 25) | θ value | 3.89403e-11 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.703954 (rank : 26) | θ value | 2.52405e-10 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.702675 (rank : 27) | θ value | 4.02038e-16 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.697045 (rank : 28) | θ value | 2.13673e-09 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.696348 (rank : 29) | θ value | 2.79066e-09 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.692033 (rank : 30) | θ value | 2.20605e-14 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.686175 (rank : 31) | θ value | 1.63604e-09 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.682380 (rank : 32) | θ value | 1.47974e-10 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.673764 (rank : 33) | θ value | 6.21693e-09 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.672450 (rank : 34) | θ value | 2.28291e-11 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.671732 (rank : 35) | θ value | 5.08577e-11 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIO_HUMAN
|
||||||
| NC score | 0.671390 (rank : 36) | θ value | 8.67504e-11 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.671214 (rank : 37) | θ value | 2.52405e-10 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.668006 (rank : 38) | θ value | 1.58096e-12 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.666313 (rank : 39) | θ value | 1.17247e-07 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.664448 (rank : 40) | θ value | 1.69304e-06 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.663085 (rank : 41) | θ value | 3.89403e-11 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.644116 (rank : 42) | θ value | 1.53129e-07 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.628752 (rank : 43) | θ value | 7.84624e-12 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.628032 (rank : 44) | θ value | 4.59992e-12 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.626390 (rank : 45) | θ value | 3.41135e-07 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.624974 (rank : 46) | θ value | 2.52405e-10 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.617356 (rank : 47) | θ value | 9.92553e-07 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.608821 (rank : 48) | θ value | 1.29631e-06 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.600771 (rank : 49) | θ value | 2.44474e-05 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.599229 (rank : 50) | θ value | 3.29651e-10 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.593873 (rank : 51) | θ value | 1.47974e-10 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.580952 (rank : 52) | θ value | 1.9326e-10 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.577395 (rank : 53) | θ value | 4.30538e-10 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.569126 (rank : 54) | θ value | 2.52405e-10 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.499829 (rank : 55) | θ value | 6.87365e-08 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.479565 (rank : 56) | θ value | 1.53129e-07 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.413900 (rank : 57) | θ value | 2.7842e-17 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.408863 (rank : 58) | θ value | 1.63225e-17 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.313786 (rank : 59) | θ value | 0.00869519 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.313309 (rank : 60) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.301146 (rank : 61) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
OCRL_HUMAN
|
||||||
| NC score | 0.284740 (rank : 62) | θ value | 0.0431538 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
PCTL_MOUSE
|
||||||
| NC score | 0.100455 (rank : 63) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PCTL_HUMAN
|
||||||
| NC score | 0.099299 (rank : 64) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
FA13A_MOUSE
|
||||||
| NC score | 0.078884 (rank : 65) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.078579 (rank : 66) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.072167 (rank : 67) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.067284 (rank : 68) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.066946 (rank : 69) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.066336 (rank : 70) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
GRP3_HUMAN
|
||||||
| NC score | 0.064409 (rank : 71) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.061349 (rank : 72) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FACD2_HUMAN
|
||||||
| NC score | 0.061017 (rank : 73) | θ value | 0.125558 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXW9, Q2LA86, Q69YP9, Q6PJN7, Q9BQ06, Q9H9T9 | Gene names | FANCD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein (Protein FACD2). | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.059688 (rank : 74) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
BNIP2_HUMAN
|
||||||
| NC score | 0.055629 (rank : 75) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12982 | Gene names | BNIP2, NIP2 | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
BNIP2_MOUSE
|
||||||
| NC score | 0.055547 (rank : 76) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54940 | Gene names | Bnip2, Nip2l | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.054290 (rank : 77) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.053707 (rank : 78) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
ATCAY_MOUSE
|
||||||
| NC score | 0.052907 (rank : 79) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin. | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.052452 (rank : 80) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
ATCAY_HUMAN
|
||||||
| NC score | 0.052424 (rank : 81) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WG3, Q8NAQ2, Q8TAQ3, Q96HC6, Q96JF5 | Gene names | ATCAY, KIAA1872 | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin (Ataxia Cayman type protein) (BNIP-H). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.051920 (rank : 82) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.051817 (rank : 83) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.051599 (rank : 84) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
BNIPL_MOUSE
|
||||||
| NC score | 0.051399 (rank : 85) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JU7, Q923E3 | Gene names | Bnipl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.051381 (rank : 86) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
BNIPL_HUMAN
|
||||||
| NC score | 0.051209 (rank : 87) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.050813 (rank : 88) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.050266 (rank : 89) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.050017 (rank : 90) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CENPQ_HUMAN
|
||||||
| NC score | 0.042372 (rank : 91) | θ value | 0.21417 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7L2Z9, Q6IN61, Q9NVS5 | Gene names | CENPQ, C6orf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.036897 (rank : 92) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.036596 (rank : 93) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.036581 (rank : 94) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
WBP4_HUMAN
|
||||||
| NC score | 0.035709 (rank : 95) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.034917 (rank : 96) | θ value | 0.00509761 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.034884 (rank : 97) | θ value | 0.00509761 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.032216 (rank : 98) | θ value | 0.21417 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.028728 (rank : 99) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.028621 (rank : 100) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SLBP_MOUSE
|
||||||
| NC score | 0.027323 (rank : 101) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97440 | Gene names | Slbp, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.027218 (rank : 102) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.026465 (rank : 103) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
SLBP_HUMAN
|
||||||
| NC score | 0.024234 (rank : 104) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14493 | Gene names | SLBP, HBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.022359 (rank : 105) | θ value | 0.47712 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
UT14B_MOUSE
|
||||||
| NC score | 0.020822 (rank : 106) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
XPO1_MOUSE
|
||||||
| NC score | 0.019977 (rank : 107) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5F9 | Gene names | Xpo1, Crm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-1 (Exp1) (Chromosome region maintenance 1 protein homolog). | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.016154 (rank : 108) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.016141 (rank : 109) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.015767 (rank : 110) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.015669 (rank : 111) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.015434 (rank : 112) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
RAE2_HUMAN
|
||||||
| NC score | 0.014270 (rank : 113) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26374, Q9H1Y4 | Gene names | CHML, REP2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.014132 (rank : 114) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.013998 (rank : 115) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
GP73_MOUSE
|
||||||
| NC score | 0.013387 (rank : 116) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.013156 (rank : 117) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
MCM4_MOUSE
|
||||||
| NC score | 0.012613 (rank : 118) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.012556 (rank : 119) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.012003 (rank : 120) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.011628 (rank : 121) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.011491 (rank : 122) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.010850 (rank : 123) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.010691 (rank : 124) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.009678 (rank : 125) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
VPS54_MOUSE
|
||||||
| NC score | 0.009032 (rank : 126) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SPW0, Q8BPB3, Q8CFZ7, Q8CHL5, Q8R3R4, Q8R3X1 | Gene names | Vps54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p homolog). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.008549 (rank : 127) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.008266 (rank : 128) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.007987 (rank : 129) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
CCND3_HUMAN
|
||||||
| NC score | 0.004546 (rank : 130) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.003661 (rank : 131) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
LAMB3_HUMAN
|
||||||
| NC score | 0.001820 (rank : 132) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
KLHL6_MOUSE
|
||||||
| NC score | 0.001261 (rank : 133) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6V595 | Gene names | Klhl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 6. | |||||
|
ATS2_MOUSE
|
||||||
| NC score | 0.001171 (rank : 134) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
NRK_MOUSE
|
||||||
| NC score | -0.004421 (rank : 135) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||
|
DAPK3_MOUSE
|
||||||
| NC score | -0.005536 (rank : 136) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54784 | Gene names | Dapk3, Zipk | |||
|
Domain Architecture |

|
|||||
| Description | Death-associated protein kinase 3 (EC 2.7.11.1) (DAP kinase 3) (DAP- like kinase) (Dlk) (ZIP-kinase). | |||||