Please be patient as the page loads
|
FA13C_HUMAN
|
||||||
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FA13C_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.964783 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13A_HUMAN
|
||||||
| θ value | 3.59435e-57 (rank : 3) | NC score | 0.840726 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
FA13A_MOUSE
|
||||||
| θ value | 3.974e-56 (rank : 4) | NC score | 0.843498 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 5.02714e-51 (rank : 5) | NC score | 0.620106 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CE005_HUMAN
|
||||||
| θ value | 1.28137e-46 (rank : 6) | NC score | 0.616766 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 7) | NC score | 0.030157 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
NRBF2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 8) | NC score | 0.072518 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.055200 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CN037_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.066184 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.049062 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SYAP1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.064712 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D5V6, Q3UI67, Q9D870 | Gene names | Syap1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapse-associated protein 1. | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.027220 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
CCD16_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.051831 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
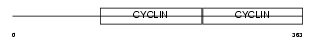
CCNE1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.034491 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.053763 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.046656 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.049687 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.021444 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.048583 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CO027_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.051303 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2M3C6, Q8N993, Q96LL5 | Gene names | C15orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.055377 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.014379 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PAK1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.011437 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13153, O75561, Q13567, Q32M53, Q32M54, Q86W79 | Gene names | PAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.047768 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.032855 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.014054 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PAK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.011356 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88643 | Gene names | Pak1, Paka | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK) (CDC42/RAC effector kinase PAK-A). | |||||
|
TMM25_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.044434 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YD3, Q6UW89, Q86UA7, Q96KA6, Q96MW9 | Gene names | TMEM25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 25 precursor. | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.036815 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.026757 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.027123 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.017871 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
HEM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.023322 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VC19, Q64453 | Gene names | Alas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.012952 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
TRI25_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.015471 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.041212 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.047302 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.012603 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.046976 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYBB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.029268 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.033076 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.029370 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
K1949_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.030571 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.021863 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.005946 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
FANCE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.036991 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB96, Q4ZGH2 | Gene names | FANCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group E protein (Protein FACE). | |||||
|
IF39_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.039757 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
JHD1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.014238 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.026639 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MDM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.024060 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00987, Q13226, Q13297, Q13298, Q13299, Q13300, Q13301, Q53XW0, Q71TW9, Q9UGI3, Q9UMT8 | Gene names | MDM2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein) (Hdm2). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.009649 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.013054 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.006087 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CP053_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.040949 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTK6 | Gene names | C16orf53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.033744 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.023415 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.013227 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.016831 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.034304 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
DNL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.021164 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
RSPO4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.013347 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.019824 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.058330 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.009647 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.015079 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
ESCO2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.015429 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q56NI9 | Gene names | ESCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.016476 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PACE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.015658 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBQ7, Q6NY01, Q8BQC9, Q8BRJ1 | Gene names | Scyl3, Pace1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-associating with the carboxyl-terminal domain of ezrin (Ezrin- binding protein PACE-1) (SCY1-like protein 3). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.027296 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.024425 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.011281 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
TLE3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.008970 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
XYLT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.012880 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
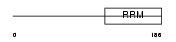
A2BP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.008354 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWB1, Q9NS20 | Gene names | A2BP1, A2BP | |||
|
Domain Architecture |
|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.027320 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.004665 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.023873 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.030972 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MPP10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.020732 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.024708 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.012126 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.013611 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.017678 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
CJ092_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.016206 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.006918 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.020731 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.023440 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.022095 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.028671 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PCDGK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.003135 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||
|
TLE3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.007624 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.009267 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013191 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
GMIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.059093 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.060249 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.051460 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
K1688_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050572 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052198 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.058259 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.070301 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.053137 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.053202 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.058898 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058358 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.053274 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.055869 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051033 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.061349 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.067597 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.067914 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052154 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.964783 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13A_MOUSE
|
||||||
| NC score | 0.843498 (rank : 3) | θ value | 3.974e-56 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.840726 (rank : 4) | θ value | 3.59435e-57 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.620106 (rank : 5) | θ value | 5.02714e-51 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.616766 (rank : 6) | θ value | 1.28137e-46 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
NRBF2_MOUSE
|
||||||
| NC score | 0.072518 (rank : 7) | θ value | 0.0193708 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.070301 (rank : 8) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.067914 (rank : 9) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.067597 (rank : 10) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.066184 (rank : 11) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
SYAP1_MOUSE
|
||||||
| NC score | 0.064712 (rank : 12) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D5V6, Q3UI67, Q9D870 | Gene names | Syap1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapse-associated protein 1. | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.061349 (rank : 13) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.060249 (rank : 14) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.059093 (rank : 15) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.058898 (rank : 16) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.058358 (rank : 17) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.058330 (rank : 18) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.058259 (rank : 19) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.055869 (rank : 20) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.055377 (rank : 21) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.055200 (rank : 22) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.053763 (rank : 23) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.053274 (rank : 24) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.053202 (rank : 25) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.053137 (rank : 26) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.052198 (rank : 27) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.052154 (rank : 28) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CCD16_HUMAN
|
||||||
| NC score | 0.051831 (rank : 29) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.051460 (rank : 30) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
CO027_HUMAN
|
||||||
| NC score | 0.051303 (rank : 31) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2M3C6, Q8N993, Q96LL5 | Gene names | C15orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27. | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.051033 (rank : 32) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.050572 (rank : 33) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.049687 (rank : 34) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.049062 (rank : 35) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.048583 (rank : 36) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.047768 (rank : 37) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.047302 (rank : 38) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.046976 (rank : 39) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.046656 (rank : 40) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TMM25_HUMAN
|
||||||
| NC score | 0.044434 (rank : 41) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YD3, Q6UW89, Q86UA7, Q96KA6, Q96MW9 | Gene names | TMEM25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 25 precursor. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.041212 (rank : 42) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CP053_HUMAN
|
||||||
| NC score | 0.040949 (rank : 43) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTK6 | Gene names | C16orf53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53. | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.039757 (rank : 44) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
FANCE_HUMAN
|
||||||
| NC score | 0.036991 (rank : 45) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB96, Q4ZGH2 | Gene names | FANCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group E protein (Protein FACE). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.036815 (rank : 46) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
CCNE1_MOUSE
|
||||||
| NC score | 0.034491 (rank : 47) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.034304 (rank : 48) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.033744 (rank : 49) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.033076 (rank : 50) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.032855 (rank : 51) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.030972 (rank : 52) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
K1949_MOUSE
|
||||||
| NC score | 0.030571 (rank : 53) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.030157 (rank : 54) | θ value | 0.000461057 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.029370 (rank : 55) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MYBB_HUMAN
|
||||||
| NC score | 0.029268 (rank : 56) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.028671 (rank : 57) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.027320 (rank : 58) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.027296 (rank : 59) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.027220 (rank : 60) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.027123 (rank : 61) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.026757 (rank : 62) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.026639 (rank : 63) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.024708 (rank : 64) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.024425 (rank : 65) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
MDM2_HUMAN
|
||||||
| NC score | 0.024060 (rank : 66) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00987, Q13226, Q13297, Q13298, Q13299, Q13300, Q13301, Q53XW0, Q71TW9, Q9UGI3, Q9UMT8 | Gene names | MDM2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein) (Hdm2). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.023873 (rank : 67) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.023440 (rank : 68) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.023415 (rank : 69) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
HEM1_MOUSE
|
||||||
| NC score | 0.023322 (rank : 70) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VC19, Q64453 | Gene names | Alas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.022095 (rank : 71) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.021863 (rank : 72) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.021444 (rank : 73) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
DNL1_MOUSE
|
||||||
| NC score | 0.021164 (rank : 74) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
MPP10_HUMAN
|
||||||
| NC score | 0.020732 (rank : 75) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.020731 (rank : 76) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.019824 (rank : 77) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.017871 (rank : 78) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.017678 (rank : 79) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.016831 (rank : 80) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.016476 (rank : 81) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CJ092_HUMAN
|
||||||
| NC score | 0.016206 (rank : 82) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
PACE1_MOUSE
|
||||||
| NC score | 0.015658 (rank : 83) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBQ7, Q6NY01, Q8BQC9, Q8BRJ1 | Gene names | Scyl3, Pace1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-associating with the carboxyl-terminal domain of ezrin (Ezrin- binding protein PACE-1) (SCY1-like protein 3). | |||||
|
TRI25_HUMAN
|
||||||
| NC score | 0.015471 (rank : 84) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
ESCO2_HUMAN
|
||||||
| NC score | 0.015429 (rank : 85) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q56NI9 | Gene names | ESCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.015079 (rank : 86) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.014379 (rank : 87) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
JHD1B_HUMAN
|
||||||
| NC score | 0.014238 (rank : 88) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.014054 (rank : 89) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.013611 (rank : 90) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
RSPO4_HUMAN
|
||||||
| NC score | 0.013347 (rank : 91) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.013227 (rank : 92) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.013191 (rank : 93) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.013054 (rank : 94) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.012952 (rank : 95) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
XYLT1_HUMAN
|
||||||
| NC score | 0.012880 (rank : 96) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.012603 (rank : 97) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.012126 (rank : 98) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
PAK1_HUMAN
|
||||||
| NC score | 0.011437 (rank : 99) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13153, O75561, Q13567, Q32M53, Q32M54, Q86W79 | Gene names | PAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK). | |||||
|
PAK1_MOUSE
|
||||||
| NC score | 0.011356 (rank : 100) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88643 | Gene names | Pak1, Paka | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK) (CDC42/RAC effector kinase PAK-A). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.011281 (rank : 101) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.009649 (rank : 102) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.009647 (rank : 103) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.009267 (rank : 104) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
TLE3_HUMAN
|
||||||
| NC score | 0.008970 (rank : 105) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
A2BP1_HUMAN
|
||||||
| NC score | 0.008354 (rank : 106) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWB1, Q9NS20 | Gene names | A2BP1, A2BP | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
TLE3_MOUSE
|
||||||
| NC score | 0.007624 (rank : 107) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.006918 (rank : 108) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.006087 (rank : 109) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.005946 (rank : 110) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.004665 (rank : 111) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
PCDGK_HUMAN
|
||||||
| NC score | 0.003135 (rank : 112) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||