Please be patient as the page loads
|
DNL1_MOUSE
|
||||||
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DNL1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.940695 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL3_MOUSE
|
||||||
| θ value | 2.50074e-42 (rank : 3) | NC score | 0.739839 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 3.26607e-42 (rank : 4) | NC score | 0.740098 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL4_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 5) | NC score | 0.683828 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BTF7, Q3UG76 | Gene names | Lig4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
DNL4_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 6) | NC score | 0.679907 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49917, Q8IY66, Q8TEU5 | Gene names | LIG4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 7) | NC score | 0.191886 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 8) | NC score | 0.143005 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 9) | NC score | 0.083881 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 10) | NC score | 0.093194 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 11) | NC score | 0.098285 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 12) | NC score | 0.053432 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 13) | NC score | 0.102882 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 14) | NC score | 0.127153 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.122934 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.073855 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.053198 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.128461 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.060225 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.151772 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.119977 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.017027 (rank : 135) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.075994 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.078546 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.157030 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.108614 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.082675 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.120618 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.099103 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.120206 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.075963 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
OSBL5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.032800 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.103171 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.119186 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
41_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.017698 (rank : 134) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.027342 (rank : 118) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.090075 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.020604 (rank : 128) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.045712 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.094662 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ZFP37_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.001976 (rank : 158) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17141, Q62514 | Gene names | Zfp37, Zfp-37 | |||
|
Domain Architecture |
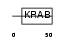
|
|||||
| Description | Zinc finger protein 37 (Zfp-37) (Male germ cell-specific zinc finger protein). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.051561 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.074270 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.132628 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.075123 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.109731 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.023322 (rank : 123) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ABCF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.033574 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.027507 (rank : 117) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.016264 (rank : 136) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
COPB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.031265 (rank : 113) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.031242 (rank : 114) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.019115 (rank : 131) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.073190 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.050913 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.069544 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.068985 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.047912 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TM115_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.026375 (rank : 119) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUH1, Q91VT6 | Gene names | Tmem115, Pl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 115 (Protein PL6 homolog). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.015438 (rank : 139) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
BCL6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.002607 (rank : 157) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |
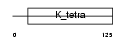
|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.020640 (rank : 127) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
FA13C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.021164 (rank : 126) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.019220 (rank : 130) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.053066 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MNDA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.033329 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |
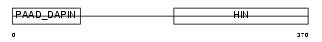
|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.018248 (rank : 133) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TLR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.006555 (rank : 152) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUN7, Q3U400, Q9DBC4 | Gene names | Tlr2 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 2 precursor (CD282 antigen). | |||||
|
ASPH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.028015 (rank : 116) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.061358 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.024192 (rank : 121) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CXAR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.016220 (rank : 137) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97792, O09052, Q3ULD0, Q91W66, Q99KG0, Q9DBJ8 | Gene names | Cxadr, Car | |||
|
Domain Architecture |
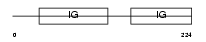
|
|||||
| Description | Coxsackievirus and adenovirus receptor homolog precursor (CAR) (mCAR). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.031363 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.039830 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KCAB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.012447 (rank : 142) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97382 | Gene names | Kcnab3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-gated potassium channel subunit beta-3 (K(+) channel subunit beta-3) (Kv-beta-3). | |||||
|
KLD7B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.010153 (rank : 146) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96G42 | Gene names | KLHDC7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 7B. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.025428 (rank : 120) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.010348 (rank : 145) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.082608 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
S26A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.009252 (rank : 147) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40879 | Gene names | SLC26A3, DRA | |||
|
Domain Architecture |

|
|||||
| Description | Chloride anion exchanger (Protein DRA) (Down-regulated in adenoma) (Solute carrier family 26 member 3). | |||||
|
TAF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.021406 (rank : 125) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.032196 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ZFP91_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.005029 (rank : 156) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.052314 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CASC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.019939 (rank : 129) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
CXAR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.015385 (rank : 140) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78310, O00694, Q8WWT6, Q8WWT7, Q8WWT8, Q9UKV4 | Gene names | CXADR, CAR | |||
|
Domain Architecture |
|
|||||
| Description | Coxsackievirus and adenovirus receptor precursor (Coxsackievirus B- adenovirus receptor) (hCAR) (CVB3-binding protein) (HCVADR). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.007300 (rank : 151) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
IF16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.033711 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.061879 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.029017 (rank : 115) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.007336 (rank : 150) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NDF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.008602 (rank : 149) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.065167 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.034824 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
SAPS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.014534 (rank : 141) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
SASH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.022744 (rank : 124) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.065421 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ZN217_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.005745 (rank : 155) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.006276 (rank : 154) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.092963 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.047262 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LOXL4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.009143 (rank : 148) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.070610 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.012406 (rank : 144) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.077678 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TAF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.018827 (rank : 132) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.016177 (rank : 138) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TRUB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.012441 (rank : 143) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWH5, Q53ES2 | Gene names | TRUB1, PUS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tRNA pseudouridine synthase 1 (EC 5.4.99.-). | |||||
|
ZFP91_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.006301 (rank : 153) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.023822 (rank : 122) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
ACINU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.057323 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.054355 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
AF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.067028 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.058038 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.058912 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.058645 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.086586 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.082868 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.093766 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CJ012_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.066377 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.053734 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.073011 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.051787 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053948 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.051745 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.055775 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056409 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
ICAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058457 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.063849 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.051654 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.058144 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.068417 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MINT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.063528 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.052900 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.056191 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
NBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.063600 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.056454 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.054349 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.051353 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.057022 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.058230 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.053025 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.068414 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.056329 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.053158 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.062559 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.064774 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.078431 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.084718 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.079454 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.077688 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.056766 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.067897 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.076606 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.057944 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.058994 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.059561 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.061313 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
DNL1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.940695 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.740098 (rank : 3) | θ value | 3.26607e-42 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_MOUSE
|
||||||
| NC score | 0.739839 (rank : 4) | θ value | 2.50074e-42 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL4_MOUSE
|
||||||
| NC score | 0.683828 (rank : 5) | θ value | 2.59387e-31 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BTF7, Q3UG76 | Gene names | Lig4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
DNL4_HUMAN
|
||||||
| NC score | 0.679907 (rank : 6) | θ value | 3.17079e-29 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49917, Q8IY66, Q8TEU5 | Gene names | LIG4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.191886 (rank : 7) | θ value | 0.000786445 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.157030 (rank : 8) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.151772 (rank : 9) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.143005 (rank : 10) | θ value | 0.000786445 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.132628 (rank : 11) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.128461 (rank : 12) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.127153 (rank : 13) | θ value | 0.00665767 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.122934 (rank : 14) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.120618 (rank : 15) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.120206 (rank : 16) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.119977 (rank : 17) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.119186 (rank : 18) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.109731 (rank : 19) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.108614 (rank : 20) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.103171 (rank : 21) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.102882 (rank : 22) | θ value | 0.00665767 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.099103 (rank : 23) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.098285 (rank : 24) | θ value | 0.00390308 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.094662 (rank : 25) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.093766 (rank : 26) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.093194 (rank : 27) | θ value | 0.00175202 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.092963 (rank : 28) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.090075 (rank : 29) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.086586 (rank : 30) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.084718 (rank : 31) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.083881 (rank : 32) | θ value | 0.00102713 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.082868 (rank : 33) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.082675 (rank : 34) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.082608 (rank : 35) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.079454 (rank : 36) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.078546 (rank : 37) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.078431 (rank : 38) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.077688 (rank : 39) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.077678 (rank : 40) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.076606 (rank : 41) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.075994 (rank : 42) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.075963 (rank : 43) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.075123 (rank : 44) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.074270 (rank : 45) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.073855 (rank : 46) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.073190 (rank : 47) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.073011 (rank : 48) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.070610 (rank : 49) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.069544 (rank : 50) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.068985 (rank : 51) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.068417 (rank : 52) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.068414 (rank : 53) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.067897 (rank : 54) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.067028 (rank : 55) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.066377 (rank : 56) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.065421 (rank : 57) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.065167 (rank : 58) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.064774 (rank : 59) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.063849 (rank : 60) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NBN_MOUSE
|
||||||
| NC score | 0.063600 (rank : 61) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.063528 (rank : 62) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.062559 (rank : 63) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.061879 (rank : 64) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.061358 (rank : 65) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.061313 (rank : 66) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.060225 (rank : 67) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.059561 (rank : 68) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.058994 (rank : 69) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.058912 (rank : 70) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.058645 (rank : 71) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.058457 (rank : 72) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.058230 (rank : 73) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.058144 (rank : 74) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.058038 (rank : 75) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.057944 (rank : 76) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.057323 (rank : 77) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.057022 (rank : 78) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.056766 (rank : 79) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.056454 (rank : 80) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.056409 (rank : 81) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.056329 (rank : 82) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
NBN_HUMAN
|
||||||
| NC score | 0.056191 (rank : 83) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.055775 (rank : 84) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.054355 (rank : 85) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.054349 (rank : 86) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.053948 (rank : 87) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.053734 (rank : 88) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.053432 (rank : 89) | θ value | 0.00665767 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.053198 (rank : 90) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.053158 (rank : 91) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.053066 (rank : 92) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.053025 (rank : 93) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.052900 (rank : 94) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.052314 (rank : 95) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.051787 (rank : 96) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.051745 (rank : 97) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.051654 (rank : 98) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.051561 (rank : 99) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.051353 (rank : 100) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.050913 (rank : 101) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.047912 (rank : 102) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.047262 (rank : 103) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.045712 (rank : 104) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.039830 (rank : 105) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.034824 (rank : 106) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.033711 (rank : 107) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
ABCF1_MOUSE
|
||||||
| NC score | 0.033574 (rank : 108) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
MNDA_HUMAN
|
||||||
| NC score | 0.033329 (rank : 109) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
OSBL5_HUMAN
|
||||||
| NC score | 0.032800 (rank : 110) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.032196 (rank : 111) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.031363 (rank : 112) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
COPB_HUMAN
|
||||||
| NC score | 0.031265 (rank : 113) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| NC score | 0.031242 (rank : 114) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.029017 (rank : 115) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.028015 (rank : 116) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.027507 (rank : 117) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.027342 (rank : 118) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
TM115_MOUSE
|
||||||
| NC score | 0.026375 (rank : 119) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUH1, Q91VT6 | Gene names | Tmem115, Pl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 115 (Protein PL6 homolog). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.025428 (rank : 120) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.024192 (rank : 121) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.023822 (rank : 122) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.023322 (rank : 123) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
SASH1_HUMAN
|
||||||
| NC score | 0.022744 (rank : 124) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
TAF2_MOUSE
|
||||||
| NC score | 0.021406 (rank : 125) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.021164 (rank : 126) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.020640 (rank : 127) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.020604 (rank : 128) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
CASC5_HUMAN
|
||||||
| NC score | 0.019939 (rank : 129) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.019220 (rank : 130) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.019115 (rank : 131) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
TAF2_HUMAN
|
||||||
| NC score | 0.018827 (rank : 132) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.018248 (rank : 133) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.017698 (rank : 134) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.017027 (rank : 135) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.016264 (rank : 136) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CXAR_MOUSE
|
||||||
| NC score | 0.016220 (rank : 137) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97792, O09052, Q3ULD0, Q91W66, Q99KG0, Q9DBJ8 | Gene names | Cxadr, Car | |||
|
Domain Architecture |

|
|||||
| Description | Coxsackievirus and adenovirus receptor homolog precursor (CAR) (mCAR). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.016177 (rank : 138) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.015438 (rank : 139) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CXAR_HUMAN
|
||||||
| NC score | 0.015385 (rank : 140) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78310, O00694, Q8WWT6, Q8WWT7, Q8WWT8, Q9UKV4 | Gene names | CXADR, CAR | |||
|
Domain Architecture |

|
|||||
| Description | Coxsackievirus and adenovirus receptor precursor (Coxsackievirus B- adenovirus receptor) (hCAR) (CVB3-binding protein) (HCVADR). | |||||
|
SAPS3_MOUSE
|
||||||
| NC score | 0.014534 (rank : 141) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
KCAB3_MOUSE
|
||||||
| NC score | 0.012447 (rank : 142) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97382 | Gene names | Kcnab3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-gated potassium channel subunit beta-3 (K(+) channel subunit beta-3) (Kv-beta-3). | |||||
|
TRUB1_HUMAN
|
||||||
| NC score | 0.012441 (rank : 143) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWH5, Q53ES2 | Gene names | TRUB1, PUS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tRNA pseudouridine synthase 1 (EC 5.4.99.-). | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.012406 (rank : 144) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.010348 (rank : 145) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
KLD7B_HUMAN
|
||||||
| NC score | 0.010153 (rank : 146) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96G42 | Gene names | KLHDC7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 7B. | |||||
|
S26A3_HUMAN
|
||||||
| NC score | 0.009252 (rank : 147) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40879 | Gene names | SLC26A3, DRA | |||
|
Domain Architecture |

|
|||||
| Description | Chloride anion exchanger (Protein DRA) (Down-regulated in adenoma) (Solute carrier family 26 member 3). | |||||
|
LOXL4_MOUSE
|
||||||
| NC score | 0.009143 (rank : 148) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
NDF6_HUMAN
|
||||||
| NC score | 0.008602 (rank : 149) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.007336 (rank : 150) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.007300 (rank : 151) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
TLR2_MOUSE
|
||||||
| NC score | 0.006555 (rank : 152) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUN7, Q3U400, Q9DBC4 | Gene names | Tlr2 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 2 precursor (CD282 antigen). | |||||
|
ZFP91_HUMAN
|
||||||
| NC score | 0.006301 (rank : 153) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.006276 (rank : 154) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
ZN217_HUMAN
|
||||||
| NC score | 0.005745 (rank : 155) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
ZFP91_MOUSE
|
||||||
| NC score | 0.005029 (rank : 156) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
BCL6_MOUSE
|
||||||
| NC score | 0.002607 (rank : 157) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
ZFP37_MOUSE
|
||||||
| NC score | 0.001976 (rank : 158) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17141, Q62514 | Gene names | Zfp37, Zfp-37 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 37 (Zfp-37) (Male germ cell-specific zinc finger protein). | |||||