Please be patient as the page loads
|
NAL10_HUMAN
|
||||||
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NAL10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988227 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 6.27148e-94 (rank : 3) | NC score | 0.921601 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 1.70789e-91 (rank : 4) | NC score | 0.912633 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 3.93732e-88 (rank : 5) | NC score | 0.917871 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 6) | NC score | 0.880529 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 1.66577e-62 (rank : 7) | NC score | 0.934738 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 2.84138e-62 (rank : 8) | NC score | 0.890562 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 2.48917e-58 (rank : 9) | NC score | 0.929837 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 6.77864e-56 (rank : 10) | NC score | 0.898128 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 6.34462e-54 (rank : 11) | NC score | 0.903836 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 5.93839e-52 (rank : 12) | NC score | 0.891652 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 13) | NC score | 0.876589 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 3.26607e-42 (rank : 14) | NC score | 0.857869 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 15) | NC score | 0.799676 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 16) | NC score | 0.848394 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 17) | NC score | 0.709627 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 18) | NC score | 0.690103 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 19) | NC score | 0.619116 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 20) | NC score | 0.645095 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CARD4_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 21) | NC score | 0.633771 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 22) | NC score | 0.637299 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 23) | NC score | 0.123837 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 24) | NC score | 0.226271 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
ASC_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 25) | NC score | 0.395278 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
BIR1A_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 26) | NC score | 0.269998 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 27) | NC score | 0.250395 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
ASC_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 28) | NC score | 0.338784 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 29) | NC score | 0.262720 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 30) | NC score | 0.286000 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
CAR12_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 31) | NC score | 0.346102 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 32) | NC score | 0.255063 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 33) | NC score | 0.259407 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
CF182_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 34) | NC score | 0.030818 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYX8 | Gene names | C6orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182. | |||||
|
PTH2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.048432 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3E5, Q9NTE5 | Gene names | PTRH2, BIT1, PTH2 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-tRNA hydrolase 2, mitochondrial precursor (EC 3.1.1.29) (PTH 2) (Bcl-2 inhibitor of transcription 1). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.013965 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.014060 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.014731 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
GP107_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.013704 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUV8, Q6ZPL4, Q8BM58, Q8BMN6, Q8BTW1 | Gene names | Gpr107, Kiaa1624 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR107 precursor. | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.016033 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.001414 (rank : 73) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.004831 (rank : 71) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CF152_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.005192 (rank : 70) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.000533 (rank : 75) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DNL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.007336 (rank : 67) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
GP73_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.008238 (rank : 66) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBJ4, Q6IAF4, Q9NRB9 | Gene names | GOLPH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
LYAR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.009795 (rank : 65) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.006553 (rank : 68) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PTH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.028802 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2Y8, Q8BI01, Q8BI31 | Gene names | Ptrh2, Pth2 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-tRNA hydrolase 2, mitochondrial precursor (EC 3.1.1.29) (PTH 2). | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.002979 (rank : 72) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.000589 (rank : 74) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GSLG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.011161 (rank : 63) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
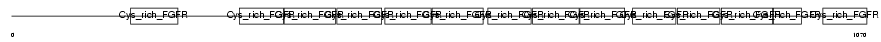
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
GSLG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.011070 (rank : 64) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
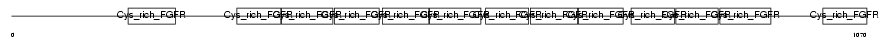
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.006391 (rank : 69) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.015042 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.058542 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.059772 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.053780 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.052598 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.054950 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |
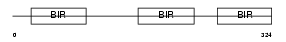
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.056075 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |
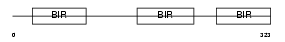
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.052991 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |
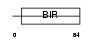
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
BIRC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.052052 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
CARD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.131657 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
CF154_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.340382 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRC14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.062280 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
LRC31_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.359318 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
LRC34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.390464 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.350133 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.114225 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.120949 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
RGP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.237737 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.227101 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RINI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.657515 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.651755 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.988227 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.934738 (rank : 3) | θ value | 1.66577e-62 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.929837 (rank : 4) | θ value | 2.48917e-58 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.921601 (rank : 5) | θ value | 6.27148e-94 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.917871 (rank : 6) | θ value | 3.93732e-88 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.912633 (rank : 7) | θ value | 1.70789e-91 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.903836 (rank : 8) | θ value | 6.34462e-54 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.898128 (rank : 9) | θ value | 6.77864e-56 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.891652 (rank : 10) | θ value | 5.93839e-52 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.890562 (rank : 11) | θ value | 2.84138e-62 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.880529 (rank : 12) | θ value | 5.17823e-64 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.876589 (rank : 13) | θ value | 1.32601e-43 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.857869 (rank : 14) | θ value | 3.26607e-42 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.848394 (rank : 15) | θ value | 2.19584e-30 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.799676 (rank : 16) | θ value | 1.62093e-41 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.709627 (rank : 17) | θ value | 7.0802e-21 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.690103 (rank : 18) | θ value | 2.69047e-20 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.657515 (rank : 19) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.651755 (rank : 20) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.645095 (rank : 21) | θ value | 8.38298e-14 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.637299 (rank : 22) | θ value | 4.76016e-09 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.633771 (rank : 23) | θ value | 1.74796e-11 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.619116 (rank : 24) | θ value | 4.91457e-14 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
ASC_MOUSE
|
||||||
| NC score | 0.395278 (rank : 25) | θ value | 6.43352e-06 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.390464 (rank : 26) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.359318 (rank : 27) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.350133 (rank : 28) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.346102 (rank : 29) | θ value | 0.000121331 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.340382 (rank : 30) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
ASC_HUMAN
|
||||||
| NC score | 0.338784 (rank : 31) | θ value | 9.29e-05 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.286000 (rank : 32) | θ value | 0.000121331 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.269998 (rank : 33) | θ value | 2.44474e-05 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.262720 (rank : 34) | θ value | 0.000121331 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.259407 (rank : 35) | θ value | 0.00175202 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.255063 (rank : 36) | θ value | 0.00020696 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.250395 (rank : 37) | θ value | 3.19293e-05 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.237737 (rank : 38) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.227101 (rank : 39) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.226271 (rank : 40) | θ value | 3.08544e-08 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
CARD8_HUMAN
|
||||||
| NC score | 0.131657 (rank : 41) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.123837 (rank : 42) | θ value | 3.08544e-08 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.120949 (rank : 43) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.114225 (rank : 44) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC14_HUMAN
|
||||||
| NC score | 0.062280 (rank : 45) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
BIRC2_MOUSE
|
||||||
| NC score | 0.059772 (rank : 46) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.058542 (rank : 47) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC4_MOUSE
|
||||||
| NC score | 0.056075 (rank : 48) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC4_HUMAN
|
||||||
| NC score | 0.054950 (rank : 49) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC3_HUMAN
|
||||||
| NC score | 0.053780 (rank : 50) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC5_HUMAN
|
||||||
| NC score | 0.052991 (rank : 51) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
BIRC3_MOUSE
|
||||||
| NC score | 0.052598 (rank : 52) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC5_MOUSE
|
||||||
| NC score | 0.052052 (rank : 53) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
PTH2_HUMAN
|
||||||
| NC score | 0.048432 (rank : 54) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3E5, Q9NTE5 | Gene names | PTRH2, BIT1, PTH2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-tRNA hydrolase 2, mitochondrial precursor (EC 3.1.1.29) (PTH 2) (Bcl-2 inhibitor of transcription 1). | |||||
|
CF182_HUMAN
|
||||||
| NC score | 0.030818 (rank : 55) | θ value | 0.0113563 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYX8 | Gene names | C6orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182. | |||||
|
PTH2_MOUSE
|
||||||
| NC score | 0.028802 (rank : 56) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2Y8, Q8BI01, Q8BI31 | Gene names | Ptrh2, Pth2 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-tRNA hydrolase 2, mitochondrial precursor (EC 3.1.1.29) (PTH 2). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.016033 (rank : 57) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.015042 (rank : 58) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.014731 (rank : 59) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.014060 (rank : 60) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.013965 (rank : 61) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
GP107_MOUSE
|
||||||
| NC score | 0.013704 (rank : 62) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUV8, Q6ZPL4, Q8BM58, Q8BMN6, Q8BTW1 | Gene names | Gpr107, Kiaa1624 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR107 precursor. | |||||
|
GSLG1_HUMAN
|
||||||
| NC score | 0.011161 (rank : 63) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
GSLG1_MOUSE
|
||||||
| NC score | 0.011070 (rank : 64) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
LYAR_MOUSE
|
||||||
| NC score | 0.009795 (rank : 65) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
GP73_HUMAN
|
||||||
| NC score | 0.008238 (rank : 66) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBJ4, Q6IAF4, Q9NRB9 | Gene names | GOLPH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
DNL1_MOUSE
|
||||||
| NC score | 0.007336 (rank : 67) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.006553 (rank : 68) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.006391 (rank : 69) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.005192 (rank : 70) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.004831 (rank : 71) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.002979 (rank : 72) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.001414 (rank : 73) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.000589 (rank : 74) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.000533 (rank : 75) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||