Please be patient as the page loads
|
NALP5_HUMAN
|
||||||
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NAL14_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.977508 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.924537 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 1.09426e-130 (rank : 4) | NC score | 0.964601 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 1.86652e-130 (rank : 5) | NC score | 0.960895 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 4.5974e-129 (rank : 6) | NC score | 0.965515 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 4.30305e-127 (rank : 7) | NC score | 0.961732 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 2.44341e-122 (rank : 8) | NC score | 0.960073 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 4.16784e-122 (rank : 9) | NC score | 0.958051 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 2.37764e-101 (rank : 10) | NC score | 0.950418 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | 7.15887e-98 (rank : 11) | NC score | 0.956804 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 5.50685e-82 (rank : 12) | NC score | 0.903474 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
RINI_MOUSE
|
||||||
| θ value | 1.15359e-63 (rank : 13) | NC score | 0.902914 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 2.17557e-62 (rank : 14) | NC score | 0.908555 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 2.65945e-60 (rank : 15) | NC score | 0.909460 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
RINI_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 16) | NC score | 0.902444 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 3.26607e-42 (rank : 17) | NC score | 0.857869 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 18) | NC score | 0.848272 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 19) | NC score | 0.695862 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
CARD4_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 20) | NC score | 0.693637 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 21) | NC score | 0.699622 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 22) | NC score | 0.715037 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 23) | NC score | 0.582852 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC34_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 24) | NC score | 0.559115 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC31_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 25) | NC score | 0.519704 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 26) | NC score | 0.180892 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CF154_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 27) | NC score | 0.548693 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 28) | NC score | 0.191109 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 29) | NC score | 0.361468 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
LRC34_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 30) | NC score | 0.520988 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
RGP1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 31) | NC score | 0.376609 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 32) | NC score | 0.586439 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CAR12_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 33) | NC score | 0.296866 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 34) | NC score | 0.152588 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MAPE_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 35) | NC score | 0.119952 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
PRA12_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 36) | NC score | 0.094622 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member DJ1198H6.2. | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 37) | NC score | 0.085553 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.032884 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
PRAM4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 39) | NC score | 0.085560 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60810, Q5LJB5 | Gene names | PRAMEF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 4. | |||||
|
PRAM5_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 40) | NC score | 0.088148 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TYX0 | Gene names | PRAMEF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 5. | |||||
|
PRAM6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 41) | NC score | 0.088155 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VXH4 | Gene names | PRAMEF6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 6. | |||||
|
PRAM9_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.084616 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VWM5 | Gene names | PRAMEF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 9. | |||||
|
TBCEL_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.054500 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5QJ74 | Gene names | LRRC35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone cofactor E-like protein (Leucine-rich repeat-containing protein 35). | |||||
|
FXL13_HUMAN
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.047327 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEE6, Q6UVW7, Q6UVW8, Q75MN5, Q86UJ5, Q8N7Y4, Q8TCL2, Q8WUF9, Q8WUG0 | Gene names | FBXL13, FBL13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 13 (F-box and leucine-rich repeat protein 13). | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.176138 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
CD14_MOUSE
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.074814 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |
|
|||||
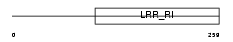
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
PRAM7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.081981 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VXH5 | Gene names | PRAMEF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 7. | |||||
|
PRAM8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.083701 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VWM4 | Gene names | PRAMEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 8. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.006267 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FXL14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.043725 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1E6 | Gene names | FBXL14, FBL14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
FXL14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.043606 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BID8, Q3U2Q9, Q8R5H7, Q8VDT7, Q922N5 | Gene names | Fbxl14, Fbl14, Ppa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
TBCEL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.038284 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5W3, Q8C7A5 | Gene names | Lrrc35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone cofactor E-like protein (Leucine-rich repeat-containing protein 35). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | -0.001640 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | -0.002303 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
LRC14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.145115 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
BRCA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.009687 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
FLII_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.015323 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JJ28, Q8K095, Q8VI44 | Gene names | Flii, Fli1, Fliih | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein flightless-1 homolog. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.011924 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LRC8D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.017575 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.020550 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.007829 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
FBXL7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.021225 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJT9, O94926 | Gene names | FBXL7, FBL6, FBL7, KIAA0840 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 7 (F-box and leucine-rich repeat protein 7) (F-box protein FBL6/FBL7). | |||||
|
LRC46_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.008734 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96FV0 | Gene names | LRRC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 46. | |||||
|
PHLPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.016542 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
BIR1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.158046 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.139293 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.151173 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.152371 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
CDY1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.007251 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CPN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.008277 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBB9, Q8R113 | Gene names | Cpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N subunit 2 precursor (Carboxypeptidase N polypeptide 2) (Carboxypeptidase N 83 kDa chain) (Carboxypeptidase N regulatory subunit) (Carboxypeptidase N large subunit). | |||||
|
FLII_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.011916 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
KCT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.017543 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K201, Q5SVC5, Q922L7, Q9CVN1 | Gene names | Kct2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
KIME_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.009463 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03426 | Gene names | MVK | |||
|
Domain Architecture |

|
|||||
| Description | Mevalonate kinase (EC 2.7.1.36) (MK). | |||||
|
KSR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | -0.002269 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.006592 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
PHLPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.014993 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
RAD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.001201 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55042, Q96F39 | Gene names | RRAD, RAD | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein RAD (RAS associated with diabetes) (RAD1). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | -0.003767 (rank : 94) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TDRD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.006783 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.144401 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
CO3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.003326 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.003517 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PRAM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.067426 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60811 | Gene names | PRAMEF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 2. | |||||
|
REM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.000855 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEL9, Q8BPB1 | Gene names | Rem2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein REM 2 (Rad and Gem-like GTP-binding protein 2). | |||||
|
TAIP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.007402 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TLR13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.015668 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
TMOD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.045701 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JHJ0, Q3TIT1 | Gene names | Tmod3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.007381 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ASC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.196834 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |
|
|||||
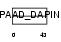
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
ASC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.249063 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
CARD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.114955 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
PRAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.065559 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95521, Q9UQP2 | Gene names | PRAMEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 1. | |||||
|
PRAM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.071466 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TYW8 | Gene names | PRAMEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 3. | |||||
|
TMOD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.050107 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZR1 | Gene names | TMOD2, NTMOD | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.977508 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.965515 (rank : 3) | θ value | 4.5974e-129 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.964601 (rank : 4) | θ value | 1.09426e-130 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.961732 (rank : 5) | θ value | 4.30305e-127 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.960895 (rank : 6) | θ value | 1.86652e-130 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.960073 (rank : 7) | θ value | 2.44341e-122 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.958051 (rank : 8) | θ value | 4.16784e-122 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.956804 (rank : 9) | θ value | 7.15887e-98 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.950418 (rank : 10) | θ value | 2.37764e-101 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.924537 (rank : 11) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.909460 (rank : 12) | θ value | 2.65945e-60 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.908555 (rank : 13) | θ value | 2.17557e-62 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.903474 (rank : 14) | θ value | 5.50685e-82 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.902914 (rank : 15) | θ value | 1.15359e-63 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.902444 (rank : 16) | θ value | 8.55514e-59 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.857869 (rank : 17) | θ value | 3.26607e-42 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.848272 (rank : 18) | θ value | 5.77852e-31 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.715037 (rank : 19) | θ value | 4.02038e-16 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.699622 (rank : 20) | θ value | 1.38178e-16 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.695862 (rank : 21) | θ value | 2.06002e-20 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.693637 (rank : 22) | θ value | 1.47631e-18 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.586439 (rank : 23) | θ value | 0.000461057 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.582852 (rank : 24) | θ value | 2.69671e-12 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.559115 (rank : 25) | θ value | 4.0297e-08 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.548693 (rank : 26) | θ value | 2.88788e-06 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.520988 (rank : 27) | θ value | 0.000158464 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.519704 (rank : 28) | θ value | 5.81887e-07 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.376609 (rank : 29) | θ value | 0.000158464 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.361468 (rank : 30) | θ value | 2.44474e-05 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.296866 (rank : 31) | θ value | 0.00175202 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
ASC_MOUSE
|
||||||
| NC score | 0.249063 (rank : 32) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
ASC_HUMAN
|
||||||
| NC score | 0.196834 (rank : 33) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.191109 (rank : 34) | θ value | 1.09739e-05 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.180892 (rank : 35) | θ value | 9.92553e-07 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.176138 (rank : 36) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.158046 (rank : 37) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.152588 (rank : 38) | θ value | 0.00665767 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.152371 (rank : 39) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.151173 (rank : 40) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
LRC14_HUMAN
|
||||||
| NC score | 0.145115 (rank : 41) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.144401 (rank : 42) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.139293 (rank : 43) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
MAPE_HUMAN
|
||||||
| NC score | 0.119952 (rank : 44) | θ value | 0.0113563 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
CARD8_HUMAN
|
||||||
| NC score | 0.114955 (rank : 45) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
PRA12_HUMAN
|
||||||
| NC score | 0.094622 (rank : 46) | θ value | 0.0113563 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member DJ1198H6.2. | |||||
|
PRAM6_HUMAN
|
||||||
| NC score | 0.088155 (rank : 47) | θ value | 0.0431538 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VXH4 | Gene names | PRAMEF6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 6. | |||||
|
PRAM5_HUMAN
|
||||||
| NC score | 0.088148 (rank : 48) | θ value | 0.0431538 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TYX0 | Gene names | PRAMEF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 5. | |||||
|
PRAM4_HUMAN
|
||||||
| NC score | 0.085560 (rank : 49) | θ value | 0.0431538 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60810, Q5LJB5 | Gene names | PRAMEF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 4. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.085553 (rank : 50) | θ value | 0.0148317 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PRAM9_HUMAN
|
||||||
| NC score | 0.084616 (rank : 51) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VWM5 | Gene names | PRAMEF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 9. | |||||
|
PRAM8_HUMAN
|
||||||
| NC score | 0.083701 (rank : 52) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VWM4 | Gene names | PRAMEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 8. | |||||
|
PRAM7_HUMAN
|
||||||
| NC score | 0.081981 (rank : 53) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VXH5 | Gene names | PRAMEF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 7. | |||||
|
CD14_MOUSE
|
||||||
| NC score | 0.074814 (rank : 54) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |

|
|||||
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
PRAM3_HUMAN
|
||||||
| NC score | 0.071466 (rank : 55) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TYW8 | Gene names | PRAMEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 3. | |||||
|
PRAM2_HUMAN
|
||||||
| NC score | 0.067426 (rank : 56) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60811 | Gene names | PRAMEF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 2. | |||||
|
PRAM1_HUMAN
|
||||||
| NC score | 0.065559 (rank : 57) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95521, Q9UQP2 | Gene names | PRAMEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 1. | |||||
|
TBCEL_HUMAN
|
||||||
| NC score | 0.054500 (rank : 58) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5QJ74 | Gene names | LRRC35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone cofactor E-like protein (Leucine-rich repeat-containing protein 35). | |||||
|
TMOD2_HUMAN
|
||||||
| NC score | 0.050107 (rank : 59) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZR1 | Gene names | TMOD2, NTMOD | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
FXL13_HUMAN
|
||||||
| NC score | 0.047327 (rank : 60) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEE6, Q6UVW7, Q6UVW8, Q75MN5, Q86UJ5, Q8N7Y4, Q8TCL2, Q8WUF9, Q8WUG0 | Gene names | FBXL13, FBL13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 13 (F-box and leucine-rich repeat protein 13). | |||||
|
TMOD3_MOUSE
|
||||||
| NC score | 0.045701 (rank : 61) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JHJ0, Q3TIT1 | Gene names | Tmod3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
FXL14_HUMAN
|
||||||
| NC score | 0.043725 (rank : 62) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1E6 | Gene names | FBXL14, FBL14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
FXL14_MOUSE
|
||||||
| NC score | 0.043606 (rank : 63) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BID8, Q3U2Q9, Q8R5H7, Q8VDT7, Q922N5 | Gene names | Fbxl14, Fbl14, Ppa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
TBCEL_MOUSE
|
||||||
| NC score | 0.038284 (rank : 64) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5W3, Q8C7A5 | Gene names | Lrrc35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone cofactor E-like protein (Leucine-rich repeat-containing protein 35). | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.032884 (rank : 65) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
FBXL7_HUMAN
|
||||||
| NC score | 0.021225 (rank : 66) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJT9, O94926 | Gene names | FBXL7, FBL6, FBL7, KIAA0840 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 7 (F-box and leucine-rich repeat protein 7) (F-box protein FBL6/FBL7). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.020550 (rank : 67) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
LRC8D_HUMAN
|
||||||
| NC score | 0.017575 (rank : 68) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
KCT2_MOUSE
|
||||||
| NC score | 0.017543 (rank : 69) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K201, Q5SVC5, Q922L7, Q9CVN1 | Gene names | Kct2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
PHLPL_HUMAN
|
||||||
| NC score | 0.016542 (rank : 70) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
TLR13_MOUSE
|
||||||
| NC score | 0.015668 (rank : 71) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
FLII_MOUSE
|
||||||
| NC score | 0.015323 (rank : 72) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JJ28, Q8K095, Q8VI44 | Gene names | Flii, Fli1, Fliih | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein flightless-1 homolog. | |||||
|
PHLPL_MOUSE
|
||||||
| NC score | 0.014993 (rank : 73) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.011924 (rank : 74) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
FLII_HUMAN
|
||||||
| NC score | 0.011916 (rank : 75) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
BRCA2_MOUSE
|
||||||
| NC score | 0.009687 (rank : 76) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
KIME_HUMAN
|
||||||
| NC score | 0.009463 (rank : 77) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03426 | Gene names | MVK | |||
|
Domain Architecture |

|
|||||
| Description | Mevalonate kinase (EC 2.7.1.36) (MK). | |||||
|
LRC46_HUMAN
|
||||||
| NC score | 0.008734 (rank : 78) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96FV0 | Gene names | LRRC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 46. | |||||
|
CPN2_MOUSE
|
||||||
| NC score | 0.008277 (rank : 79) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBB9, Q8R113 | Gene names | Cpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N subunit 2 precursor (Carboxypeptidase N polypeptide 2) (Carboxypeptidase N 83 kDa chain) (Carboxypeptidase N regulatory subunit) (Carboxypeptidase N large subunit). | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.007829 (rank : 80) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
TAIP2_MOUSE
|
||||||
| NC score | 0.007402 (rank : 81) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.007381 (rank : 82) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.007251 (rank : 83) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
TDRD1_MOUSE
|
||||||
| NC score | 0.006783 (rank : 84) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.006592 (rank : 85) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.006267 (rank : 86) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.003517 (rank : 87) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
CO3_HUMAN
|
||||||
| NC score | 0.003326 (rank : 88) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
RAD_HUMAN
|
||||||
| NC score | 0.001201 (rank : 89) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55042, Q96F39 | Gene names | RRAD, RAD | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein RAD (RAS associated with diabetes) (RAD1). | |||||
|
REM2_MOUSE
|
||||||
| NC score | 0.000855 (rank : 90) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEL9, Q8BPB1 | Gene names | Rem2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein REM 2 (Rad and Gem-like GTP-binding protein 2). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.001640 (rank : 91) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
KSR1_HUMAN
|
||||||
| NC score | -0.002269 (rank : 92) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | -0.002303 (rank : 93) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
STK10_MOUSE
|
||||||
| NC score | -0.003767 (rank : 94) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||