Please be patient as the page loads
|
NAL12_HUMAN
|
||||||
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CIAS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987696 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987031 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 4.53388e-177 (rank : 4) | NC score | 0.976793 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 2.19974e-139 (rank : 5) | NC score | 0.973527 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 3.51195e-137 (rank : 6) | NC score | 0.966900 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 4.58675e-137 (rank : 7) | NC score | 0.969654 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 1.86652e-130 (rank : 8) | NC score | 0.960895 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 5.08302e-128 (rank : 9) | NC score | 0.927796 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 4.7686e-118 (rank : 10) | NC score | 0.892308 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 3.0981e-109 (rank : 11) | NC score | 0.946103 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 3.42535e-108 (rank : 12) | NC score | 0.960325 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | 1.06727e-101 (rank : 13) | NC score | 0.947435 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 1.88391e-98 (rank : 14) | NC score | 0.944673 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 1.70789e-91 (rank : 15) | NC score | 0.912633 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 1.95861e-79 (rank : 16) | NC score | 0.907035 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
RINI_HUMAN
|
||||||
| θ value | 5.51964e-74 (rank : 17) | NC score | 0.886462 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| θ value | 1.40689e-69 (rank : 18) | NC score | 0.883237 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
CARD4_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 19) | NC score | 0.732129 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 20) | NC score | 0.771422 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 21) | NC score | 0.757196 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 22) | NC score | 0.729326 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 23) | NC score | 0.656124 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 24) | NC score | 0.631808 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC31_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 25) | NC score | 0.537650 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 26) | NC score | 0.186891 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC34_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 27) | NC score | 0.552785 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 28) | NC score | 0.254762 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
LRC34_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 29) | NC score | 0.517239 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
BIR1A_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 30) | NC score | 0.238020 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 31) | NC score | 0.101264 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 32) | NC score | 0.183664 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 33) | NC score | 0.230241 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 34) | NC score | 0.359134 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 35) | NC score | 0.172915 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
RGP1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 36) | NC score | 0.376079 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 37) | NC score | 0.218828 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 38) | NC score | 0.231388 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 39) | NC score | 0.223549 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
CAR12_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 40) | NC score | 0.356346 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
CD14_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 41) | NC score | 0.081628 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |
|
|||||
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
CF154_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 42) | NC score | 0.499814 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRRC1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 43) | NC score | 0.048391 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 44) | NC score | 0.057847 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
LRC8D_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.048958 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
LRCH4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.042348 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
LRRC1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 47) | NC score | 0.047143 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 48) | NC score | 0.060330 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
LRC15_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 49) | NC score | 0.039382 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80X72 | Gene names | Lrrc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 15 precursor. | |||||
|
LRC15_HUMAN
|
||||||
| θ value | 0.125558 (rank : 50) | NC score | 0.034111 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TF66, Q7RTN7 | Gene names | LRRC15, LIB | |||
|
Domain Architecture |
|
|||||
| Description | Leucine-rich repeat-containing protein 15 precursor (hLib). | |||||
|
FXL14_HUMAN
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.042969 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1E6 | Gene names | FBXL14, FBL14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
FXL14_MOUSE
|
||||||
| θ value | 0.163984 (rank : 52) | NC score | 0.042845 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BID8, Q3U2Q9, Q8R5H7, Q8VDT7, Q922N5 | Gene names | Fbxl14, Fbl14, Ppa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
SHC3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.018485 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92529, Q8TAP2, Q9UCX5 | Gene names | SHC3, NSHC, SHCC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc) (Protein Rai). | |||||
|
XPO2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 54) | NC score | 0.031996 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55060, O75432, Q32M40, Q9H5B7, Q9NTS0, Q9UP98, Q9UP99, Q9UPA0 | Gene names | CSE1L, CAS, XPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-2 (Exp2) (Importin-alpha re-exporter) (Chromosome segregation 1-like protein) (Cellular apoptosis susceptibility protein). | |||||
|
XPO2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 55) | NC score | 0.031981 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERK4 | Gene names | Cse1l, Xpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-2 (Exp2) (Importin-alpha re-exporter) (Chromosome segregation 1-like protein). | |||||
|
LRC8D_MOUSE
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.044727 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
PRAM3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.023252 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TYW8 | Gene names | PRAMEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 3. | |||||
|
ASC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.301480 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.034597 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.038454 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
SHC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.016623 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61120 | Gene names | Shc3, Nshc, ShcC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc). | |||||
|
ASC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.248113 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
LRC33_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.039069 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86YC3 | Gene names | LRRC33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 33 precursor. | |||||
|
LRC8E_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.041646 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q66JT1 | Gene names | Lrrc8e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
FLII_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.039741 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJ28, Q8K095, Q8VI44 | Gene names | Flii, Fli1, Fliih | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein flightless-1 homolog. | |||||
|
TLR13_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.033971 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
AASS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.014432 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99K67, Q9Z1I9 | Gene names | Aass, Lorsdh | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
AIM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.045968 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
LRC57_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.049296 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D1G5, Q9DC95 | Gene names | Lrrc57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 57. | |||||
|
PHLPL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.048471 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
CPN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.028752 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBB9, Q8R113 | Gene names | Cpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N subunit 2 precursor (Carboxypeptidase N polypeptide 2) (Carboxypeptidase N 83 kDa chain) (Carboxypeptidase N regulatory subunit) (Carboxypeptidase N large subunit). | |||||
|
LRC47_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.041905 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N1G4, Q9ULN5 | Gene names | LRRC47, KIAA1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
PHLPL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.049056 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PIDD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.039972 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HB75, Q59FD1, Q59H10, Q59HC7, Q7Z4P8, Q8NC89, Q8NDL2, Q96C25, Q9NRE6 | Gene names | LRDD, PIDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein (p53-induced protein with a death domain). | |||||
|
TLR11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.030949 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6R5P0 | Gene names | Tlr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 11 precursor. | |||||
|
TLR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.022375 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15455 | Gene names | TLR3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 3 precursor (CD283 antigen). | |||||
|
LRC39_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.038951 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGI7 | Gene names | Lrrc39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 39. | |||||
|
LRCH4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.035285 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
MAPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.059432 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
TMOD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.046737 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYL9, Q9NT43, Q9NZR0 | Gene names | TMOD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
CCG8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.007227 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXS5, Q9BXT0, Q9BY23 | Gene names | CACNG8, CACNG6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
ELMO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.008189 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92556, Q29R79, Q96PB0, Q9H0I1 | Gene names | ELMO1, KIAA0281 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
ELMO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.008196 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BPU7, Q8BSY9, Q8K2C5, Q91ZU3 | Gene names | Elmo1 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
FLII_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.037302 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
LRC14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.095974 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
NPHP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.011960 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59240 | Gene names | Nphp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
CCG8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.006650 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHW2 | Gene names | Cacng8 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
DPP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.007618 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NY33, O95748, Q969H2, Q9BV67, Q9HAL6 | Gene names | DPP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 3 (EC 3.4.14.4) (Dipeptidyl-peptidase III) (DPP III) (Dipeptidyl aminopeptidase III) (Dipeptidyl arylamidase III). | |||||
|
LRC33_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.036890 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BMT4, Q3TIA8, Q8BTT4, Q8BUI7, Q8BY16, Q8R063 | Gene names | Lrrc33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 33 precursor. | |||||
|
TMOD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.053228 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHJ0, Q3TIT1 | Gene names | Tmod3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
DIRA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.006099 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |
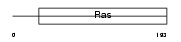
|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
DNM1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.003882 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
LRIG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.015198 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6UXM1, Q6UXL7, Q8NC72 | Gene names | LRIG3, LIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 3 precursor (LIG-3). | |||||
|
NDUB9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.007858 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6M9, Q9UQE8 | Gene names | NDUFB9, UQOR22 | |||
|
Domain Architecture |
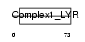
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B22 subunit) (Complex I-B22) (CI-B22). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.004252 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.006688 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
CARD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.124460 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
TMOD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.059623 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZR1 | Gene names | TMOD2, NTMOD | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
TMOD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.050729 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKK7, Q9JLH9 | Gene names | Tmod2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
TMOD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.052721 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZQ9, Q8WVL3, Q9UKH2 | Gene names | TMOD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-4 (Skeletal muscle tropomodulin) (Sk-Tmod). | |||||
|
TMOD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.050384 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLH8 | Gene names | Tmod4 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-4 (Skeletal muscle tropomodulin) (Sk-Tmod). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.987696 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.987031 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.976793 (rank : 4) | θ value | 4.53388e-177 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.973527 (rank : 5) | θ value | 2.19974e-139 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.969654 (rank : 6) | θ value | 4.58675e-137 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.966900 (rank : 7) | θ value | 3.51195e-137 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.960895 (rank : 8) | θ value | 1.86652e-130 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.960325 (rank : 9) | θ value | 3.42535e-108 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.947435 (rank : 10) | θ value | 1.06727e-101 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.946103 (rank : 11) | θ value | 3.0981e-109 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.944673 (rank : 12) | θ value | 1.88391e-98 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.927796 (rank : 13) | θ value | 5.08302e-128 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.912633 (rank : 14) | θ value | 1.70789e-91 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.907035 (rank : 15) | θ value | 1.95861e-79 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.892308 (rank : 16) | θ value | 4.7686e-118 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.886462 (rank : 17) | θ value | 5.51964e-74 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.883237 (rank : 18) | θ value | 1.40689e-69 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.771422 (rank : 19) | θ value | 6.82597e-32 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.757196 (rank : 20) | θ value | 1.52067e-31 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.732129 (rank : 21) | θ value | 1.0531e-32 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.729326 (rank : 22) | θ value | 5.97985e-28 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.656124 (rank : 23) | θ value | 7.32683e-18 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.631808 (rank : 24) | θ value | 3.63628e-17 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.552785 (rank : 25) | θ value | 9.92553e-07 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.537650 (rank : 26) | θ value | 1.33837e-11 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.517239 (rank : 27) | θ value | 2.88788e-06 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.499814 (rank : 28) | θ value | 0.0193708 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.376079 (rank : 29) | θ value | 0.000121331 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.359134 (rank : 30) | θ value | 3.19293e-05 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.356346 (rank : 31) | θ value | 0.00509761 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
ASC_MOUSE
|
||||||
| NC score | 0.301480 (rank : 32) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.254762 (rank : 33) | θ value | 1.29631e-06 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
ASC_HUMAN
|
||||||
| NC score | 0.248113 (rank : 34) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.238020 (rank : 35) | θ value | 1.09739e-05 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.231388 (rank : 36) | θ value | 0.00175202 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.230241 (rank : 37) | θ value | 2.44474e-05 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.223549 (rank : 38) | θ value | 0.00228821 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.218828 (rank : 39) | θ value | 0.00035302 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.186891 (rank : 40) | θ value | 2.36244e-08 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.183664 (rank : 41) | θ value | 1.87187e-05 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.172915 (rank : 42) | θ value | 4.1701e-05 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CARD8_HUMAN
|
||||||
| NC score | 0.124460 (rank : 43) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.101264 (rank : 44) | θ value | 1.09739e-05 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC14_HUMAN
|
||||||
| NC score | 0.095974 (rank : 45) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
CD14_MOUSE
|
||||||
| NC score | 0.081628 (rank : 46) | θ value | 0.00869519 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |

|
|||||
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.060330 (rank : 47) | θ value | 0.0736092 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
TMOD2_HUMAN
|
||||||
| NC score | 0.059623 (rank : 48) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZR1 | Gene names | TMOD2, NTMOD | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
MAPE_HUMAN
|
||||||
| NC score | 0.059432 (rank : 49) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.057847 (rank : 50) | θ value | 0.0193708 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
TMOD3_MOUSE
|
||||||
| NC score | 0.053228 (rank : 51) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHJ0, Q3TIT1 | Gene names | Tmod3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
TMOD4_HUMAN
|
||||||
| NC score | 0.052721 (rank : 52) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZQ9, Q8WVL3, Q9UKH2 | Gene names | TMOD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-4 (Skeletal muscle tropomodulin) (Sk-Tmod). | |||||
|
TMOD2_MOUSE
|
||||||
| NC score | 0.050729 (rank : 53) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKK7, Q9JLH9 | Gene names | Tmod2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
TMOD4_MOUSE
|
||||||
| NC score | 0.050384 (rank : 54) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLH8 | Gene names | Tmod4 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-4 (Skeletal muscle tropomodulin) (Sk-Tmod). | |||||
|
LRC57_MOUSE
|
||||||
| NC score | 0.049296 (rank : 55) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D1G5, Q9DC95 | Gene names | Lrrc57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 57. | |||||
|
PHLPL_HUMAN
|
||||||
| NC score | 0.049056 (rank : 56) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
LRC8D_HUMAN
|
||||||
| NC score | 0.048958 (rank : 57) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7L1W4, Q6UWB2, Q9NVW3 | Gene names | LRRC8D, LRRC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
PHLPL_MOUSE
|
||||||
| NC score | 0.048471 (rank : 58) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
LRRC1_HUMAN
|
||||||
| NC score | 0.048391 (rank : 59) | θ value | 0.0193708 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
LRRC1_MOUSE
|
||||||
| NC score | 0.047143 (rank : 60) | θ value | 0.0431538 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
TMOD3_HUMAN
|
||||||
| NC score | 0.046737 (rank : 61) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYL9, Q9NT43, Q9NZR0 | Gene names | TMOD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod). | |||||
|
AIM2_HUMAN
|
||||||
| NC score | 0.045968 (rank : 62) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
LRC8D_MOUSE
|
||||||
| NC score | 0.044727 (rank : 63) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
FXL14_HUMAN
|
||||||
| NC score | 0.042969 (rank : 64) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1E6 | Gene names | FBXL14, FBL14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
FXL14_MOUSE
|
||||||
| NC score | 0.042845 (rank : 65) | θ value | 0.163984 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BID8, Q3U2Q9, Q8R5H7, Q8VDT7, Q922N5 | Gene names | Fbxl14, Fbl14, Ppa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
LRCH4_MOUSE
|
||||||
| NC score | 0.042348 (rank : 66) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
LRC47_HUMAN
|
||||||
| NC score | 0.041905 (rank : 67) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N1G4, Q9ULN5 | Gene names | LRRC47, KIAA1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
LRC8E_MOUSE
|
||||||
| NC score | 0.041646 (rank : 68) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q66JT1 | Gene names | Lrrc8e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
PIDD_HUMAN
|
||||||
| NC score | 0.039972 (rank : 69) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HB75, Q59FD1, Q59H10, Q59HC7, Q7Z4P8, Q8NC89, Q8NDL2, Q96C25, Q9NRE6 | Gene names | LRDD, PIDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein (p53-induced protein with a death domain). | |||||
|
FLII_MOUSE
|
||||||
| NC score | 0.039741 (rank : 70) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJ28, Q8K095, Q8VI44 | Gene names | Flii, Fli1, Fliih | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein flightless-1 homolog. | |||||
|
LRC15_MOUSE
|
||||||
| NC score | 0.039382 (rank : 71) | θ value | 0.0961366 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80X72 | Gene names | Lrrc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 15 precursor. | |||||
|
LRC33_HUMAN
|
||||||
| NC score | 0.039069 (rank : 72) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86YC3 | Gene names | LRRC33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 33 precursor. | |||||
|
LRC39_MOUSE
|
||||||
| NC score | 0.038951 (rank : 73) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGI7 | Gene names | Lrrc39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 39. | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.038454 (rank : 74) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
FLII_HUMAN
|
||||||
| NC score | 0.037302 (rank : 75) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
LRC33_MOUSE
|
||||||
| NC score | 0.036890 (rank : 76) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BMT4, Q3TIA8, Q8BTT4, Q8BUI7, Q8BY16, Q8R063 | Gene names | Lrrc33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 33 precursor. | |||||
|
LRCH4_HUMAN
|
||||||
| NC score | 0.035285 (rank : 77) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.034597 (rank : 78) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
LRC15_HUMAN
|
||||||
| NC score | 0.034111 (rank : 79) | θ value | 0.125558 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TF66, Q7RTN7 | Gene names | LRRC15, LIB | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 15 precursor (hLib). | |||||
|
TLR13_MOUSE
|
||||||
| NC score | 0.033971 (rank : 80) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
XPO2_HUMAN
|
||||||
| NC score | 0.031996 (rank : 81) | θ value | 0.21417 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55060, O75432, Q32M40, Q9H5B7, Q9NTS0, Q9UP98, Q9UP99, Q9UPA0 | Gene names | CSE1L, CAS, XPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-2 (Exp2) (Importin-alpha re-exporter) (Chromosome segregation 1-like protein) (Cellular apoptosis susceptibility protein). | |||||
|
XPO2_MOUSE
|
||||||
| NC score | 0.031981 (rank : 82) | θ value | 0.21417 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERK4 | Gene names | Cse1l, Xpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-2 (Exp2) (Importin-alpha re-exporter) (Chromosome segregation 1-like protein). | |||||
|
TLR11_MOUSE
|
||||||
| NC score | 0.030949 (rank : 83) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6R5P0 | Gene names | Tlr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 11 precursor. | |||||
|
CPN2_MOUSE
|
||||||
| NC score | 0.028752 (rank : 84) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBB9, Q8R113 | Gene names | Cpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N subunit 2 precursor (Carboxypeptidase N polypeptide 2) (Carboxypeptidase N 83 kDa chain) (Carboxypeptidase N regulatory subunit) (Carboxypeptidase N large subunit). | |||||
|
PRAM3_HUMAN
|
||||||
| NC score | 0.023252 (rank : 85) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TYW8 | Gene names | PRAMEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 3. | |||||
|
TLR3_HUMAN
|
||||||
| NC score | 0.022375 (rank : 86) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15455 | Gene names | TLR3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 3 precursor (CD283 antigen). | |||||
|
SHC3_HUMAN
|
||||||
| NC score | 0.018485 (rank : 87) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92529, Q8TAP2, Q9UCX5 | Gene names | SHC3, NSHC, SHCC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc) (Protein Rai). | |||||
|
SHC3_MOUSE
|
||||||
| NC score | 0.016623 (rank : 88) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61120 | Gene names | Shc3, Nshc, ShcC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc). | |||||
|
LRIG3_HUMAN
|
||||||
| NC score | 0.015198 (rank : 89) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6UXM1, Q6UXL7, Q8NC72 | Gene names | LRIG3, LIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 3 precursor (LIG-3). | |||||
|
AASS_MOUSE
|
||||||
| NC score | 0.014432 (rank : 90) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99K67, Q9Z1I9 | Gene names | Aass, Lorsdh | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
NPHP4_MOUSE
|
||||||
| NC score | 0.011960 (rank : 91) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59240 | Gene names | Nphp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
ELMO1_MOUSE
|
||||||
| NC score | 0.008196 (rank : 92) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BPU7, Q8BSY9, Q8K2C5, Q91ZU3 | Gene names | Elmo1 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
ELMO1_HUMAN
|
||||||
| NC score | 0.008189 (rank : 93) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92556, Q29R79, Q96PB0, Q9H0I1 | Gene names | ELMO1, KIAA0281 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
NDUB9_HUMAN
|
||||||
| NC score | 0.007858 (rank : 94) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6M9, Q9UQE8 | Gene names | NDUFB9, UQOR22 | |||
|
Domain Architecture |
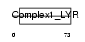
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B22 subunit) (Complex I-B22) (CI-B22). | |||||
|
DPP3_HUMAN
|
||||||
| NC score | 0.007618 (rank : 95) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NY33, O95748, Q969H2, Q9BV67, Q9HAL6 | Gene names | DPP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 3 (EC 3.4.14.4) (Dipeptidyl-peptidase III) (DPP III) (Dipeptidyl aminopeptidase III) (Dipeptidyl arylamidase III). | |||||
|
CCG8_HUMAN
|
||||||
| NC score | 0.007227 (rank : 96) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXS5, Q9BXT0, Q9BY23 | Gene names | CACNG8, CACNG6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.006688 (rank : 97) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
CCG8_MOUSE
|
||||||
| NC score | 0.006650 (rank : 98) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHW2 | Gene names | Cacng8 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
DIRA3_HUMAN
|
||||||
| NC score | 0.006099 (rank : 99) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.004252 (rank : 100) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
DNM1L_HUMAN
|
||||||
| NC score | 0.003882 (rank : 101) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||