Please be patient as the page loads
|
BIR1A_MOUSE
|
||||||
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BIR1A_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995448 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996218 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.995217 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994757 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.992756 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
CAR12_HUMAN
|
||||||
| θ value | 3.04986e-48 (rank : 7) | NC score | 0.759681 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 8) | NC score | 0.704498 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC4_MOUSE
|
||||||
| θ value | 4.13158e-37 (rank : 9) | NC score | 0.721975 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |
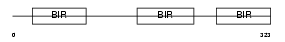
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC2_MOUSE
|
||||||
| θ value | 1.20211e-36 (rank : 10) | NC score | 0.700617 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC3_HUMAN
|
||||||
| θ value | 2.05049e-36 (rank : 11) | NC score | 0.698151 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC3_MOUSE
|
||||||
| θ value | 7.79185e-36 (rank : 12) | NC score | 0.695606 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC4_HUMAN
|
||||||
| θ value | 1.73584e-35 (rank : 13) | NC score | 0.715550 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |
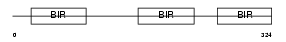
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC7_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 14) | NC score | 0.624354 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |
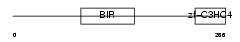
|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
BIRC8_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 15) | NC score | 0.629293 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |
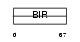
|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
BIRC5_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 16) | NC score | 0.620793 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
BIRC5_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 17) | NC score | 0.617640 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 18) | NC score | 0.281819 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 19) | NC score | 0.285176 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
LRC31_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 20) | NC score | 0.338971 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 21) | NC score | 0.250926 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 22) | NC score | 0.296275 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
BIRC6_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 23) | NC score | 0.324270 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 24) | NC score | 0.271682 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 25) | NC score | 0.238020 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 26) | NC score | 0.269998 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 27) | NC score | 0.259416 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 28) | NC score | 0.225317 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 29) | NC score | 0.239591 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 30) | NC score | 0.218889 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 31) | NC score | 0.255118 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
LRC15_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 32) | NC score | 0.054313 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80X72 | Gene names | Lrrc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 15 precursor. | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 33) | NC score | 0.260531 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 34) | NC score | 0.232340 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
TLR8_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 35) | NC score | 0.042440 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58682, Q91XI7 | Gene names | Tlr8 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 8 precursor. | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 36) | NC score | 0.196961 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
LRC18_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 37) | NC score | 0.061550 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CQ07, Q6P8T8, Q80ZT3, Q9CW01, Q9DA89 | Gene names | Lrrc18, Mtlr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 18 (Testis-specific LRR protein). | |||||
|
LRC18_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 38) | NC score | 0.053508 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N456, Q6UY02 | Gene names | LRRC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 18. | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.154083 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
FBXL7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.029083 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJT9, O94926 | Gene names | FBXL7, FBL6, FBL7, KIAA0840 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 7 (F-box and leucine-rich repeat protein 7) (F-box protein FBL6/FBL7). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.033687 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
FLII_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.044680 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.162709 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC47_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.048883 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q505F5, Q3U380, Q6ZPW2, Q9CUZ0 | Gene names | Lrrc47, Kiaa1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
GPV_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.033499 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08742 | Gene names | Gp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein V precursor (GPV) (CD42D antigen). | |||||
|
LRRC7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.037037 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80TE7 | Gene names | Lrrc7, Kiaa1365, Lap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
LRRN5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.022922 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75325, Q5T0Y0, Q6UXM0, Q8N182 | Gene names | LRRN5, GAC1, LRRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats neuronal protein 5 precursor (Glioma amplified on chromosome 1 protein). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.032640 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.004774 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CPN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.027530 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22792, Q86SU4, Q8N5V4 | Gene names | CPN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N subunit 2 precursor (Carboxypeptidase N polypeptide 2) (Carboxypeptidase N 83 kDa chain) (Carboxypeptidase N regulatory subunit) (Carboxypeptidase N large subunit). | |||||
|
LRRC7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.035884 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
PXDC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.057511 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.024193 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
OL494_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.006044 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VG07 | Gene names | Olfr494, Mor204-10 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 494 (Olfactory receptor 204-10). | |||||
|
SHOC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.039313 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
SHOC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.042249 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.012070 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PXDC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.050655 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UX71, Q96E59, Q96PD9, Q96SU9 | Gene names | PLXDC2, TEM7R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.003856 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
OL498_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.008299 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEW2 | Gene names | Olfr498, Mor204-36 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 498 (Olfactory receptor 204-36). | |||||
|
PLK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | -0.001685 (rank : 118) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07832 | Gene names | Plk1, Plk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK1 (EC 2.7.11.21) (Polo-like kinase 1) (PLK-1) (Serine/threonine-protein kinase 13) (STPK13). | |||||
|
SIX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.025566 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPC8, Q9BXH7 | Gene names | SIX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
SIX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.025200 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62232, P70179 | Gene names | Six2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
OL497_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.004868 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VG08 | Gene names | Olfr497, Mor204-9 | |||
|
Domain Architecture |
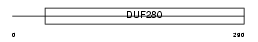
|
|||||
| Description | Olfactory receptor 497 (Olfactory receptor 204-9). | |||||
|
OL510_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.004176 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEW6 | Gene names | Olfr510, Mor204-34 | |||
|
Domain Architecture |
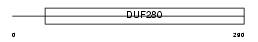
|
|||||
| Description | Olfactory receptor 510 (Olfactory receptor 204-34). | |||||
|
PLK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | -0.001861 (rank : 119) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53350, Q15153, Q99746 | Gene names | PLK1, PLK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK1 (EC 2.7.11.21) (Polo-like kinase 1) (PLK-1) (Serine/threonine-protein kinase 13) (STPK13). | |||||
|
TLR9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.048076 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQU3, Q99MF2, Q99MQ8 | Gene names | Tlr9 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 9 precursor (CD289 antigen). | |||||
|
TRIM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.009349 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||
|
TRIM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.009384 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
LRC47_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.042451 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N1G4, Q9ULN5 | Gene names | LRRC47, KIAA1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
MX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.006456 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
OL495_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.004688 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VF12 | Gene names | Olfr495, Mor204-37 | |||
|
Domain Architecture |
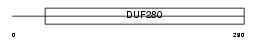
|
|||||
| Description | Olfactory receptor 495 (Olfactory receptor 204-37). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.003738 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.162669 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.016326 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
OL490_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.002043 (rank : 115) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VFD2 | Gene names | Olfr490, Mor204-17 | |||
|
Domain Architecture |
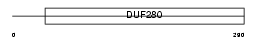
|
|||||
| Description | Olfactory receptor 490 (Olfactory receptor 204-17). | |||||
|
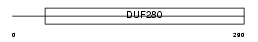
OL493_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.004120 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEW5 | Gene names | Olfr493, Mor204-35 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 493 (Olfactory receptor 204-35). | |||||
|
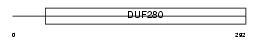
OL507_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.004256 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VG13 | Gene names | Olfr507, Mor204-7 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 507 (Olfactory receptor 204-7). | |||||
|
SIX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.020012 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15475, Q96H64 | Gene names | SIX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX1 (Sine oculis homeobox homolog 1). | |||||
|
SIX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.019999 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62231, Q8CIL7 | Gene names | Six1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX1 (Sine oculis homeobox homolog 1). | |||||
|
FLRT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.017403 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZU1 | Gene names | FLRT1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT1 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 1). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.003210 (rank : 114) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
LRC8D_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.030361 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
LSHR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.006776 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30730 | Gene names | Lhcgr, Lhr | |||
|
Domain Architecture |

|
|||||
| Description | Lutropin-choriogonadotropic hormone receptor precursor (LH/CG-R) (LSH- R) (Luteinizing hormone receptor). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.158046 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
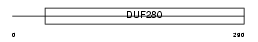
OL486_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.004766 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VFD0 | Gene names | Olfr486, Mor204-19 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 486 (Olfactory receptor 204-19). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.003563 (rank : 113) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
ABCF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.007458 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUQ8, Q86UA2, Q8NAN1, Q96GS8, Q9H7A8 | Gene names | ABCF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 3. | |||||
|
ABCF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.007256 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K268, Q9JL49 | Gene names | Abcf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 3. | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.001495 (rank : 117) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CD180_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.021362 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62192 | Gene names | Cd180, Ly78, Rp105 | |||
|
Domain Architecture |

|
|||||
| Description | CD180 antigen precursor (Lymphocyte antigen 78) (Radioprotective 105 kDa protein). | |||||
|
IF31_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.027001 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75822, Q9BUD2, Q9H8Q2, Q9UI65 | Gene names | EIF3S1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 1 (eIF-3 alpha) (eIF3 p35) (eIF3j). | |||||
|
LRRC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.058316 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
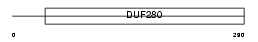
OL482_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.001713 (rank : 116) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VG03 | Gene names | Olfr482, Mor204-14 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 482 (Olfactory receptor 204-14). | |||||
|
ASC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.054705 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.103477 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CARD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.140989 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CARD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.059096 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
CF154_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059617 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
CGRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.076785 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99675, Q96BX2 | Gene names | CGRRF1, CGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
CGRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.074440 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMJ7, Q3U3F1, Q8CI24 | Gene names | Cgrrf1, Cgr19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
LRC34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.072518 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.064578 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.058701 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058677 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
LRSM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.067653 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.081091 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.067369 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.141514 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NEUL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.061484 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76050, Q5TDR2, Q5TDR3, Q8TAN0, Q9H463 | Gene names | NEURL, NEURL1, RNF67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuralized-like protein 1 (h-neuralized 1) (h-neu) (RING finger protein 67). | |||||
|
NEURL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.063627 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q923S6, Q91ZT6, Q9CWF1, Q9QWF1 | Gene names | Neurl, Neurl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuralized-like protein 1 (m-neuralized 1) (m-neu1). | |||||
|
RFFL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.110027 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
RFFL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.111915 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
RINI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.125545 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.122638 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.084046 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
RNF26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.124760 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BY78 | Gene names | RNF26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 26. | |||||
|
RNF34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.109875 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
RNF34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.110397 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.996218 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.995448 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.995217 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.994757 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.992756 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.759681 (rank : 7) | θ value | 3.04986e-48 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
BIRC4_MOUSE
|
||||||
| NC score | 0.721975 (rank : 8) | θ value | 4.13158e-37 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC4_HUMAN
|
||||||
| NC score | 0.715550 (rank : 9) | θ value | 1.73584e-35 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.704498 (rank : 10) | θ value | 4.88048e-38 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC2_MOUSE
|
||||||
| NC score | 0.700617 (rank : 11) | θ value | 1.20211e-36 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC3_HUMAN
|
||||||
| NC score | 0.698151 (rank : 12) | θ value | 2.05049e-36 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC3_MOUSE
|
||||||
| NC score | 0.695606 (rank : 13) | θ value | 7.79185e-36 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC8_HUMAN
|
||||||
| NC score | 0.629293 (rank : 14) | θ value | 8.95645e-16 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
BIRC7_HUMAN
|
||||||
| NC score | 0.624354 (rank : 15) | θ value | 7.82807e-20 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
BIRC5_MOUSE
|
||||||
| NC score | 0.620793 (rank : 16) | θ value | 1.38499e-08 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
BIRC5_HUMAN
|
||||||
| NC score | 0.617640 (rank : 17) | θ value | 2.36244e-08 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.338971 (rank : 18) | θ value | 3.41135e-07 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
BIRC6_HUMAN
|
||||||
| NC score | 0.324270 (rank : 19) | θ value | 2.88788e-06 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.296275 (rank : 20) | θ value | 1.69304e-06 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.285176 (rank : 21) | θ value | 3.41135e-07 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.281819 (rank : 22) | θ value | 8.97725e-08 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.271682 (rank : 23) | θ value | 2.88788e-06 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.269998 (rank : 24) | θ value | 2.44474e-05 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.260531 (rank : 25) | θ value | 0.00102713 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.259416 (rank : 26) | θ value | 4.1701e-05 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.255118 (rank : 27) | θ value | 0.000602161 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.250926 (rank : 28) | θ value | 3.41135e-07 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.239591 (rank : 29) | θ value | 9.29e-05 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.238020 (rank : 30) | θ value | 1.09739e-05 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.232340 (rank : 31) | θ value | 0.00390308 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.225317 (rank : 32) | θ value | 9.29e-05 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.218889 (rank : 33) | θ value | 0.000121331 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.196961 (rank : 34) | θ value | 0.0330416 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.162709 (rank : 35) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.162669 (rank : 36) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.158046 (rank : 37) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.154083 (rank : 38) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.141514 (rank : 39) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.140989 (rank : 40) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.125545 (rank : 41) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RNF26_HUMAN
|
||||||
| NC score | 0.124760 (rank : 42) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BY78 | Gene names | RNF26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 26. | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.122638 (rank : 43) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
RFFL_MOUSE
|
||||||
| NC score | 0.111915 (rank : 44) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
RNF34_MOUSE
|
||||||
| NC score | 0.110397 (rank : 45) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
RFFL_HUMAN
|
||||||
| NC score | 0.110027 (rank : 46) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
RNF34_HUMAN
|
||||||
| NC score | 0.109875 (rank : 47) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.103477 (rank : 48) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.084046 (rank : 49) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.081091 (rank : 50) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
CGRF1_HUMAN
|
||||||
| NC score | 0.076785 (rank : 51) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99675, Q96BX2 | Gene names | CGRRF1, CGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
CGRF1_MOUSE
|
||||||
| NC score | 0.074440 (rank : 52) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMJ7, Q3U3F1, Q8CI24 | Gene names | Cgrrf1, Cgr19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.072518 (rank : 53) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRSM1_MOUSE
|
||||||
| NC score | 0.067653 (rank : 54) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.067369 (rank : 55) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.064578 (rank : 56) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
NEURL_MOUSE
|
||||||
| NC score | 0.063627 (rank : 57) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q923S6, Q91ZT6, Q9CWF1, Q9QWF1 | Gene names | Neurl, Neurl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuralized-like protein 1 (m-neuralized 1) (m-neu1). | |||||
|
LRC18_MOUSE
|
||||||
| NC score | 0.061550 (rank : 58) | θ value | 0.0736092 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CQ07, Q6P8T8, Q80ZT3, Q9CW01, Q9DA89 | Gene names | Lrrc18, Mtlr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 18 (Testis-specific LRR protein). | |||||
|
NEUL1_HUMAN
|
||||||
| NC score | 0.061484 (rank : 59) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76050, Q5TDR2, Q5TDR3, Q8TAN0, Q9H463 | Gene names | NEURL, NEURL1, RNF67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuralized-like protein 1 (h-neuralized 1) (h-neu) (RING finger protein 67). | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.059617 (rank : 60) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
CARD8_HUMAN
|
||||||
| NC score | 0.059096 (rank : 61) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
LRRC1_HUMAN
|
||||||
| NC score | 0.058701 (rank : 62) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.058677 (rank : 63) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
LRRC1_MOUSE
|
||||||
| NC score | 0.058316 (rank : 64) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
PXDC2_MOUSE
|
||||||
| NC score | 0.057511 (rank : 65) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
ASC_MOUSE
|
||||||
| NC score | 0.054705 (rank : 66) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
LRC15_MOUSE
|
||||||
| NC score | 0.054313 (rank : 67) | θ value | 0.00102713 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80X72 | Gene names | Lrrc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 15 precursor. | |||||
|
LRC18_HUMAN
|
||||||
| NC score | 0.053508 (rank : 68) | θ value | 0.0961366 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N456, Q6UY02 | Gene names | LRRC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 18. | |||||
|
PXDC2_HUMAN
|
||||||
| NC score | 0.050655 (rank : 69) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UX71, Q96E59, Q96PD9, Q96SU9 | Gene names | PLXDC2, TEM7R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
LRC47_MOUSE
|
||||||
| NC score | 0.048883 (rank : 70) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q505F5, Q3U380, Q6ZPW2, Q9CUZ0 | Gene names | Lrrc47, Kiaa1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
TLR9_MOUSE
|
||||||
| NC score | 0.048076 (rank : 71) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQU3, Q99MF2, Q99MQ8 | Gene names | Tlr9 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 9 precursor (CD289 antigen). | |||||
|
FLII_HUMAN
|
||||||
| NC score | 0.044680 (rank : 72) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
LRC47_HUMAN
|
||||||
| NC score | 0.042451 (rank : 73) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N1G4, Q9ULN5 | Gene names | LRRC47, KIAA1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
TLR8_MOUSE
|
||||||
| NC score | 0.042440 (rank : 74) | θ value | 0.0252991 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58682, Q91XI7 | Gene names | Tlr8 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 8 precursor. | |||||
|
SHOC2_MOUSE
|
||||||
| NC score | 0.042249 (rank : 75) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
SHOC2_HUMAN
|
||||||
| NC score | 0.039313 (rank : 76) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
LRRC7_MOUSE
|
||||||
| NC score | 0.037037 (rank : 77) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80TE7 | Gene names | Lrrc7, Kiaa1365, Lap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
LRRC7_HUMAN
|
||||||
| NC score | 0.035884 (rank : 78) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.033687 (rank : 79) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
GPV_MOUSE
|
||||||
| NC score | 0.033499 (rank : 80) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08742 | Gene names | Gp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein V precursor (GPV) (CD42D antigen). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.032640 (rank : 81) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
LRC8D_MOUSE
|
||||||
| NC score | 0.030361 (rank : 82) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
FBXL7_HUMAN
|
||||||
| NC score | 0.029083 (rank : 83) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJT9, O94926 | Gene names | FBXL7, FBL6, FBL7, KIAA0840 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 7 (F-box and leucine-rich repeat protein 7) (F-box protein FBL6/FBL7). | |||||
|
CPN2_HUMAN
|
||||||
| NC score | 0.027530 (rank : 84) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22792, Q86SU4, Q8N5V4 | Gene names | CPN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N subunit 2 precursor (Carboxypeptidase N polypeptide 2) (Carboxypeptidase N 83 kDa chain) (Carboxypeptidase N regulatory subunit) (Carboxypeptidase N large subunit). | |||||
|
IF31_HUMAN
|
||||||
| NC score | 0.027001 (rank : 85) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75822, Q9BUD2, Q9H8Q2, Q9UI65 | Gene names | EIF3S1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 1 (eIF-3 alpha) (eIF3 p35) (eIF3j). | |||||
|
SIX2_HUMAN
|
||||||
| NC score | 0.025566 (rank : 86) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPC8, Q9BXH7 | Gene names | SIX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
SIX2_MOUSE
|
||||||
| NC score | 0.025200 (rank : 87) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62232, P70179 | Gene names | Six2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.024193 (rank : 88) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
LRRN5_HUMAN
|
||||||
| NC score | 0.022922 (rank : 89) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75325, Q5T0Y0, Q6UXM0, Q8N182 | Gene names | LRRN5, GAC1, LRRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats neuronal protein 5 precursor (Glioma amplified on chromosome 1 protein). | |||||
|
CD180_MOUSE
|
||||||
| NC score | 0.021362 (rank : 90) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62192 | Gene names | Cd180, Ly78, Rp105 | |||
|
Domain Architecture |

|
|||||
| Description | CD180 antigen precursor (Lymphocyte antigen 78) (Radioprotective 105 kDa protein). | |||||
|
SIX1_HUMAN
|
||||||
| NC score | 0.020012 (rank : 91) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15475, Q96H64 | Gene names | SIX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX1 (Sine oculis homeobox homolog 1). | |||||
|
SIX1_MOUSE
|
||||||
| NC score | 0.019999 (rank : 92) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62231, Q8CIL7 | Gene names | Six1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX1 (Sine oculis homeobox homolog 1). | |||||
|
FLRT1_HUMAN
|
||||||
| NC score | 0.017403 (rank : 93) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZU1 | Gene names | FLRT1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT1 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.016326 (rank : 94) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.012070 (rank : 95) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
TRIM2_MOUSE
|
||||||
| NC score | 0.009384 (rank : 96) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
TRIM2_HUMAN
|
||||||
| NC score | 0.009349 (rank : 97) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||
|
OL498_MOUSE
|
||||||
| NC score | 0.008299 (rank : 98) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEW2 | Gene names | Olfr498, Mor204-36 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 498 (Olfactory receptor 204-36). | |||||
|
ABCF3_HUMAN
|
||||||
| NC score | 0.007458 (rank : 99) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUQ8, Q86UA2, Q8NAN1, Q96GS8, Q9H7A8 | Gene names | ABCF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 3. | |||||
|
ABCF3_MOUSE
|
||||||
| NC score | 0.007256 (rank : 100) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K268, Q9JL49 | Gene names | Abcf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 3. | |||||
|
LSHR_MOUSE
|
||||||
| NC score | 0.006776 (rank : 101) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30730 | Gene names | Lhcgr, Lhr | |||
|
Domain Architecture |

|
|||||
| Description | Lutropin-choriogonadotropic hormone receptor precursor (LH/CG-R) (LSH- R) (Luteinizing hormone receptor). | |||||
|
MX1_MOUSE
|
||||||
| NC score | 0.006456 (rank : 102) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
OL494_MOUSE
|
||||||
| NC score | 0.006044 (rank : 103) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VG07 | Gene names | Olfr494, Mor204-10 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 494 (Olfactory receptor 204-10). | |||||
|
OL497_MOUSE
|
||||||
| NC score | 0.004868 (rank : 104) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VG08 | Gene names | Olfr497, Mor204-9 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 497 (Olfactory receptor 204-9). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.004774 (rank : 105) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
OL486_MOUSE
|
||||||
| NC score | 0.004766 (rank : 106) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VFD0 | Gene names | Olfr486, Mor204-19 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 486 (Olfactory receptor 204-19). | |||||
|
OL495_MOUSE
|
||||||
| NC score | 0.004688 (rank : 107) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VF12 | Gene names | Olfr495, Mor204-37 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 495 (Olfactory receptor 204-37). | |||||
|
OL507_MOUSE
|
||||||
| NC score | 0.004256 (rank : 108) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VG13 | Gene names | Olfr507, Mor204-7 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 507 (Olfactory receptor 204-7). | |||||
|
OL510_MOUSE
|
||||||
| NC score | 0.004176 (rank : 109) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEW6 | Gene names | Olfr510, Mor204-34 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 510 (Olfactory receptor 204-34). | |||||
|
OL493_MOUSE
|
||||||
| NC score | 0.004120 (rank : 110) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEW5 | Gene names | Olfr493, Mor204-35 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 493 (Olfactory receptor 204-35). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.003856 (rank : 111) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.003738 (rank : 112) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.003563 (rank : 113) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.003210 (rank : 114) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
OL490_MOUSE
|
||||||
| NC score | 0.002043 (rank : 115) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VFD2 | Gene names | Olfr490, Mor204-17 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 490 (Olfactory receptor 204-17). | |||||
|
OL482_MOUSE
|
||||||
| NC score | 0.001713 (rank : 116) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VG03 | Gene names | Olfr482, Mor204-14 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 482 (Olfactory receptor 204-14). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.001495 (rank : 117) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
PLK1_MOUSE
|
||||||
| NC score | -0.001685 (rank : 118) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07832 | Gene names | Plk1, Plk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK1 (EC 2.7.11.21) (Polo-like kinase 1) (PLK-1) (Serine/threonine-protein kinase 13) (STPK13). | |||||
|
PLK1_HUMAN
|
||||||
| NC score | -0.001861 (rank : 119) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53350, Q15153, Q99746 | Gene names | PLK1, PLK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK1 (EC 2.7.11.21) (Polo-like kinase 1) (PLK-1) (Serine/threonine-protein kinase 13) (STPK13). | |||||