Please be patient as the page loads
|
NALP1_HUMAN
|
||||||
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NALP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 5.08302e-128 (rank : 2) | NC score | 0.927796 (rank : 2) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 8.11517e-126 (rank : 3) | NC score | 0.925572 (rank : 3) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 3.65119e-118 (rank : 4) | NC score | 0.924306 (rank : 4) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 8.41738e-115 (rank : 5) | NC score | 0.923679 (rank : 5) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 5.14231e-88 (rank : 6) | NC score | 0.919032 (rank : 8) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 1.49618e-87 (rank : 7) | NC score | 0.919168 (rank : 7) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 6.28603e-86 (rank : 8) | NC score | 0.919256 (rank : 6) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
CARD8_HUMAN
|
||||||
| θ value | 4.21644e-82 (rank : 9) | NC score | 0.393267 (rank : 31) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 5.50685e-82 (rank : 10) | NC score | 0.903474 (rank : 12) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 3.34088e-79 (rank : 11) | NC score | 0.915250 (rank : 9) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 3.12697e-77 (rank : 12) | NC score | 0.842772 (rank : 16) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 1.22964e-73 (rank : 13) | NC score | 0.910068 (rank : 10) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 7.97034e-73 (rank : 14) | NC score | 0.906399 (rank : 11) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 15) | NC score | 0.880529 (rank : 14) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | 5.35859e-61 (rank : 16) | NC score | 0.891391 (rank : 13) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 6.34462e-54 (rank : 17) | NC score | 0.872254 (rank : 15) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
RINI_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 18) | NC score | 0.816649 (rank : 17) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 19) | NC score | 0.813230 (rank : 18) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
ASC_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 20) | NC score | 0.439888 (rank : 29) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
ASC_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 21) | NC score | 0.409423 (rank : 30) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 22) | NC score | 0.714552 (rank : 19) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 23) | NC score | 0.696370 (rank : 20) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 24) | NC score | 0.651110 (rank : 22) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 25) | NC score | 0.653337 (rank : 21) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 26) | NC score | 0.586160 (rank : 24) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 27) | NC score | 0.605764 (rank : 23) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 28) | NC score | 0.275852 (rank : 38) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
LRC31_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 29) | NC score | 0.513113 (rank : 25) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 30) | NC score | 0.297279 (rank : 35) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1A_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 31) | NC score | 0.281819 (rank : 36) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 32) | NC score | 0.276047 (rank : 37) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 33) | NC score | 0.269054 (rank : 39) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 34) | NC score | 0.263277 (rank : 40) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 35) | NC score | 0.179108 (rank : 41) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 36) | NC score | 0.169959 (rank : 42) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CF154_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 37) | NC score | 0.489583 (rank : 27) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRC14_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 38) | NC score | 0.156576 (rank : 43) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
LRC34_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 39) | NC score | 0.471373 (rank : 28) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC34_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 40) | NC score | 0.504691 (rank : 26) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
PRA12_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 41) | NC score | 0.088483 (rank : 46) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member DJ1198H6.2. | |||||
|
CAR12_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 42) | NC score | 0.381207 (rank : 32) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
MAPE_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 43) | NC score | 0.109009 (rank : 45) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
PRAM7_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 44) | NC score | 0.079190 (rank : 49) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VXH5 | Gene names | PRAMEF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 7. | |||||
|
PRAM8_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.079484 (rank : 47) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VWM4 | Gene names | PRAMEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 8. | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 46) | NC score | 0.302584 (rank : 34) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
LRC33_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 47) | NC score | 0.032817 (rank : 74) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86YC3 | Gene names | LRRC33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 33 precursor. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.067208 (rank : 55) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 49) | NC score | 0.051748 (rank : 69) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
LRC8E_MOUSE
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.033402 (rank : 73) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q66JT1 | Gene names | Lrrc8e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
LRC20_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.039337 (rank : 70) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CI70 | Gene names | Lrrc20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 20. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.279714 (rank : 52) | NC score | 0.012189 (rank : 107) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LGR5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.018646 (rank : 90) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75473, Q9UP75 | Gene names | LGR5, GPR49, GPR67 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 5 precursor (Orphan G-protein coupled receptor HG38) (G-protein coupled receptor 49) (G-protein coupled receptor 67). | |||||
|
LRC32_HUMAN
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.021837 (rank : 86) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14392, Q86V06 | Gene names | LRRC32, GARP | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 32 precursor (GARP protein) (Garpin) (Glycoprotein A repetitions predominant). | |||||
|
LRC8E_HUMAN
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.029620 (rank : 77) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NSJ5, Q2YDY3, Q7L236, Q8N3B0, Q9H5H8 | Gene names | LRRC8E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
PIDD_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.030905 (rank : 76) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HB75, Q59FD1, Q59H10, Q59HC7, Q7Z4P8, Q8NC89, Q8NDL2, Q96C25, Q9NRE6 | Gene names | LRDD, PIDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein (p53-induced protein with a death domain). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.027337 (rank : 80) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.009319 (rank : 117) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.014970 (rank : 101) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.014728 (rank : 103) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.003355 (rank : 139) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
CCD22_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.016254 (rank : 97) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIG7, Q8BYH4 | Gene names | Ccdc22, DXImx40e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 22. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.010132 (rank : 110) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | -0.000703 (rank : 150) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.018473 (rank : 92) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.009289 (rank : 118) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CEP68_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.020164 (rank : 88) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.018119 (rank : 93) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.008198 (rank : 125) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
LRC8B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.025876 (rank : 81) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P9F7, Q6UY21, Q8N106, Q92627 | Gene names | LRRC8B, KIAA0231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8B (T-cell activation leucine repeat-rich protein). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.017054 (rank : 96) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.021975 (rank : 84) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CD45_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.007260 (rank : 129) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
LRC57_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.035744 (rank : 71) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N9N7, Q7Z2Z6, Q8N1T6 | Gene names | LRRC57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 57. | |||||
|
LRC8C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.027361 (rank : 79) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R502, Q3TEP9, Q8C296, Q8R0N7, Q8R3G5 | Gene names | Lrrc8c, Ad158 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8C (Protein AD158). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | -0.001135 (rank : 151) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.014899 (rank : 102) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
KCT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.024798 (rank : 82) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K201, Q5SVC5, Q922L7, Q9CVN1 | Gene names | Kct2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
NPHP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.014450 (rank : 104) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75161 | Gene names | NPHP4, KIAA0673 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
PCD20_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.001067 (rank : 146) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.015018 (rank : 100) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.017943 (rank : 94) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TLR3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.017561 (rank : 95) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99MB1, Q91ZM4 | Gene names | Tlr3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 3 precursor (CD283 antigen). | |||||
|
TYK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | -0.003037 (rank : 155) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29597, Q6QB10, Q96CH0 | Gene names | TYK2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.007615 (rank : 128) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.006133 (rank : 134) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
LRC59_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.021892 (rank : 85) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
LRC59_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.021760 (rank : 87) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q922Q8, Q3TJ35, Q3TLC7, Q3TWT9, Q3TX86 | Gene names | Lrrc59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
LRC8C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.027667 (rank : 78) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDW0, Q9H075 | Gene names | LRRC8C, AD158 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8C (Protein AD158). | |||||
|
LRRK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.002578 (rank : 143) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q38SD2, Q6NVH5, Q6NYC0, Q6ZNL9, Q6ZNM9, Q96JN5, Q9H5S3 | Gene names | LRRK1, KIAA1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
MARK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | -0.004038 (rank : 157) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHJ5, Q69ZI7 | Gene names | Mark1, Emk3, Kiaa1477 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK1 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 1) (ELKL motif serine/threonine-protein kinase 3). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.007635 (rank : 127) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TRH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.019741 (rank : 89) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62361, Q7TMT4 | Gene names | Trh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroliberin precursor [Contains: Prothyroliberin; Thyroliberin (Thyrotropin-releasing hormone) (TRH) (Thyrotropin-releasing factor) (TRF) (TSH-releasing factor) (Protirelin)]. | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | -0.001805 (rank : 153) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.015992 (rank : 99) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.004211 (rank : 138) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.009257 (rank : 119) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GA2L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.010104 (rank : 111) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
GATA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.009111 (rank : 120) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.001815 (rank : 145) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LEG3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.008669 (rank : 123) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.009522 (rank : 115) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.005239 (rank : 136) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.005584 (rank : 135) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.016219 (rank : 98) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.008206 (rank : 124) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.018547 (rank : 91) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
LRC57_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.034339 (rank : 72) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D1G5, Q9DC95 | Gene names | Lrrc57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 57. | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.013907 (rank : 105) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.009733 (rank : 113) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
DIRA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.006199 (rank : 132) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |
|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
GATA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.007988 (rank : 126) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
KAISO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | -0.002277 (rank : 154) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BN78, Q8C226, Q9WTZ7 | Gene names | Zbtb33, Kaiso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.002877 (rank : 142) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.002975 (rank : 141) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.012674 (rank : 106) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.006559 (rank : 131) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ORC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.008789 (rank : 121) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |
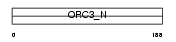
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.001827 (rank : 144) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.000053 (rank : 149) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.000457 (rank : 148) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006173 (rank : 133) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
CI079_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.009481 (rank : 116) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.004816 (rank : 137) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
EMIL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.006654 (rank : 130) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NT22, Q76KT4 | Gene names | EMILIN3, C20orf130, EMILIN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface-located protein 5) (Elastin microfibril interfacer 5). | |||||
|
F100A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.008705 (rank : 122) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TB05, Q71MF6 | Gene names | FAM100A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
GLI3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | -0.003623 (rank : 156) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.022842 (rank : 83) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.003196 (rank : 140) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.000546 (rank : 147) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
LRC20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.030921 (rank : 75) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCA0, Q5T6D4, Q5T6D6, Q9NVA6, Q9NVG3 | Gene names | LRRC20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 20. | |||||
|
MOD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.010762 (rank : 109) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |
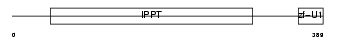
|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.009536 (rank : 114) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NTR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | -0.001558 (rank : 152) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70310 | Gene names | Ntsr2 | |||
|
Domain Architecture |
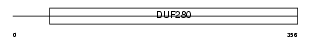
|
|||||
| Description | Neurotensin receptor type 2 (NT-R-2) (Low-affinity levocabastine- sensitive neurotensin receptor) (NTRL). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.009737 (rank : 112) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PRAM5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.067847 (rank : 53) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TYX0 | Gene names | PRAMEF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 5. | |||||
|
PRAM6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.067920 (rank : 52) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VXH4 | Gene names | PRAMEF6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 6. | |||||
|
TMM28_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.011240 (rank : 108) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.068462 (rank : 51) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.073854 (rank : 50) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.067001 (rank : 56) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.063420 (rank : 61) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.066293 (rank : 57) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |
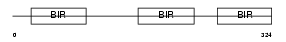
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.067683 (rank : 54) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |
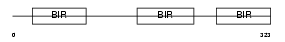
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.063879 (rank : 59) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |
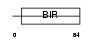
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
BIRC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.062743 (rank : 62) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |
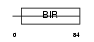
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
BIRC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.060033 (rank : 64) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |
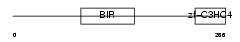
|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
BIRC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.054376 (rank : 67) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
CD14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.053685 (rank : 68) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |
|
|||||
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.079412 (rank : 48) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MEFV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.145886 (rank : 44) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PRAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.056281 (rank : 65) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95521, Q9UQP2 | Gene names | PRAMEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 1. | |||||
|
PRAM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.055394 (rank : 66) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60811 | Gene names | PRAMEF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 2. | |||||
|
PRAM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.060668 (rank : 63) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TYW8 | Gene names | PRAMEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 3. | |||||
|
PRAM4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.064236 (rank : 58) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60810, Q5LJB5 | Gene names | PRAMEF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 4. | |||||
|
PRAM9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.063485 (rank : 60) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5VWM5 | Gene names | PRAMEF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 9. | |||||
|
RGP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.302938 (rank : 33) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.927796 (rank : 2) | θ value | 5.08302e-128 (rank : 2) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.925572 (rank : 3) | θ value | 8.11517e-126 (rank : 3) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.924306 (rank : 4) | θ value | 3.65119e-118 (rank : 4) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.923679 (rank : 5) | θ value | 8.41738e-115 (rank : 5) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.919256 (rank : 6) | θ value | 6.28603e-86 (rank : 8) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.919168 (rank : 7) | θ value | 1.49618e-87 (rank : 7) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.919032 (rank : 8) | θ value | 5.14231e-88 (rank : 6) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.915250 (rank : 9) | θ value | 3.34088e-79 (rank : 11) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.910068 (rank : 10) | θ value | 1.22964e-73 (rank : 13) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.906399 (rank : 11) | θ value | 7.97034e-73 (rank : 14) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.903474 (rank : 12) | θ value | 5.50685e-82 (rank : 10) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.891391 (rank : 13) | θ value | 5.35859e-61 (rank : 16) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.880529 (rank : 14) | θ value | 5.17823e-64 (rank : 15) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.872254 (rank : 15) | θ value | 6.34462e-54 (rank : 17) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.842772 (rank : 16) | θ value | 3.12697e-77 (rank : 12) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.816649 (rank : 17) | θ value | 2.12192e-33 (rank : 18) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.813230 (rank : 18) | θ value | 2.12192e-33 (rank : 19) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.714552 (rank : 19) | θ value | 7.0802e-21 (rank : 22) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.696370 (rank : 20) | θ value | 7.82807e-20 (rank : 23) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.653337 (rank : 21) | θ value | 2.69671e-12 (rank : 25) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.651110 (rank : 22) | θ value | 1.2105e-12 (rank : 24) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.605764 (rank : 23) | θ value | 1.47974e-10 (rank : 27) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.586160 (rank : 24) | θ value | 1.02475e-11 (rank : 26) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.513113 (rank : 25) | θ value | 8.11959e-09 (rank : 29) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.504691 (rank : 26) | θ value | 0.00020696 (rank : 40) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.489583 (rank : 27) | θ value | 1.87187e-05 (rank : 37) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.471373 (rank : 28) | θ value | 7.1131e-05 (rank : 39) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
ASC_MOUSE
|
||||||
| NC score | 0.439888 (rank : 29) | θ value | 1.42661e-21 (rank : 20) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
ASC_HUMAN
|
||||||
| NC score | 0.409423 (rank : 30) | θ value | 7.0802e-21 (rank : 21) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
CARD8_HUMAN
|
||||||
| NC score | 0.393267 (rank : 31) | θ value | 4.21644e-82 (rank : 9) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.381207 (rank : 32) | θ value | 0.00102713 (rank : 42) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.302938 (rank : 33) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.302584 (rank : 34) | θ value | 0.00390308 (rank : 46) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.297279 (rank : 35) | θ value | 3.08544e-08 (rank : 30) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.281819 (rank : 36) | θ value | 8.97725e-08 (rank : 31) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.276047 (rank : 37) | θ value | 1.17247e-07 (rank : 32) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.275852 (rank : 38) | θ value | 6.21693e-09 (rank : 28) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.269054 (rank : 39) | θ value | 1.53129e-07 (rank : 33) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.263277 (rank : 40) | θ value | 1.99992e-07 (rank : 34) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.179108 (rank : 41) | θ value | 1.99992e-07 (rank : 35) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.169959 (rank : 42) | θ value | 9.92553e-07 (rank : 36) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC14_HUMAN
|
||||||
| NC score | 0.156576 (rank : 43) | θ value | 3.19293e-05 (rank : 38) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.145886 (rank : 44) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MAPE_HUMAN
|
||||||
| NC score | 0.109009 (rank : 45) | θ value | 0.00228821 (rank : 43) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
PRA12_HUMAN
|
||||||
| NC score | 0.088483 (rank : 46) | θ value | 0.000602161 (rank : 41) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member DJ1198H6.2. | |||||
|
PRAM8_HUMAN
|
||||||
| NC score | 0.079484 (rank : 47) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VWM4 | Gene names | PRAMEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 8. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.079412 (rank : 48) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PRAM7_HUMAN
|
||||||
| NC score | 0.079190 (rank : 49) | θ value | 0.00228821 (rank : 44) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VXH5 | Gene names | PRAMEF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 7. | |||||
|
BIRC2_MOUSE
|
||||||
| NC score | 0.073854 (rank : 50) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.068462 (rank : 51) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
PRAM6_HUMAN
|
||||||
| NC score | 0.067920 (rank : 52) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VXH4 | Gene names | PRAMEF6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 6. | |||||
|
PRAM5_HUMAN
|
||||||
| NC score | 0.067847 (rank : 53) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TYX0 | Gene names | PRAMEF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 5. | |||||
|
BIRC4_MOUSE
|
||||||
| NC score | 0.067683 (rank : 54) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.067208 (rank : 55) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
BIRC3_HUMAN
|
||||||
| NC score | 0.067001 (rank : 56) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC4_HUMAN
|
||||||
| NC score | 0.066293 (rank : 57) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
PRAM4_HUMAN
|
||||||
| NC score | 0.064236 (rank : 58) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60810, Q5LJB5 | Gene names | PRAMEF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 4. | |||||
|
BIRC5_HUMAN
|
||||||
| NC score | 0.063879 (rank : 59) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
PRAM9_HUMAN
|
||||||
| NC score | 0.063485 (rank : 60) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5VWM5 | Gene names | PRAMEF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 9. | |||||
|
BIRC3_MOUSE
|
||||||
| NC score | 0.063420 (rank : 61) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC5_MOUSE
|
||||||
| NC score | 0.062743 (rank : 62) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
PRAM3_HUMAN
|
||||||
| NC score | 0.060668 (rank : 63) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TYW8 | Gene names | PRAMEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 3. | |||||
|
BIRC7_HUMAN
|
||||||
| NC score | 0.060033 (rank : 64) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
PRAM1_HUMAN
|
||||||
| NC score | 0.056281 (rank : 65) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95521, Q9UQP2 | Gene names | PRAMEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 1. | |||||
|
PRAM2_HUMAN
|
||||||
| NC score | 0.055394 (rank : 66) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60811 | Gene names | PRAMEF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 2. | |||||
|
BIRC8_HUMAN
|
||||||
| NC score | 0.054376 (rank : 67) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
CD14_MOUSE
|
||||||
| NC score | 0.053685 (rank : 68) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |

|
|||||
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.051748 (rank : 69) | θ value | 0.0961366 (rank : 49) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
LRC20_MOUSE
|
||||||
| NC score | 0.039337 (rank : 70) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CI70 | Gene names | Lrrc20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 20. | |||||
|
LRC57_HUMAN
|
||||||
| NC score | 0.035744 (rank : 71) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N9N7, Q7Z2Z6, Q8N1T6 | Gene names | LRRC57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 57. | |||||
|
LRC57_MOUSE
|
||||||
| NC score | 0.034339 (rank : 72) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D1G5, Q9DC95 | Gene names | Lrrc57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 57. | |||||
|
LRC8E_MOUSE
|
||||||
| NC score | 0.033402 (rank : 73) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q66JT1 | Gene names | Lrrc8e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
LRC33_HUMAN
|
||||||
| NC score | 0.032817 (rank : 74) | θ value | 0.0431538 (rank : 47) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86YC3 | Gene names | LRRC33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 33 precursor. | |||||
|
LRC20_HUMAN
|
||||||
| NC score | 0.030921 (rank : 75) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCA0, Q5T6D4, Q5T6D6, Q9NVA6, Q9NVG3 | Gene names | LRRC20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 20. | |||||
|
PIDD_HUMAN
|
||||||
| NC score | 0.030905 (rank : 76) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HB75, Q59FD1, Q59H10, Q59HC7, Q7Z4P8, Q8NC89, Q8NDL2, Q96C25, Q9NRE6 | Gene names | LRDD, PIDD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein (p53-induced protein with a death domain). | |||||
|
LRC8E_HUMAN
|
||||||
| NC score | 0.029620 (rank : 77) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NSJ5, Q2YDY3, Q7L236, Q8N3B0, Q9H5H8 | Gene names | LRRC8E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8E. | |||||
|
LRC8C_HUMAN
|
||||||
| NC score | 0.027667 (rank : 78) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDW0, Q9H075 | Gene names | LRRC8C, AD158 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8C (Protein AD158). | |||||
|
LRC8C_MOUSE
|
||||||
| NC score | 0.027361 (rank : 79) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R502, Q3TEP9, Q8C296, Q8R0N7, Q8R3G5 | Gene names | Lrrc8c, Ad158 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8C (Protein AD158). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.027337 (rank : 80) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
LRC8B_HUMAN
|
||||||
| NC score | 0.025876 (rank : 81) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P9F7, Q6UY21, Q8N106, Q92627 | Gene names | LRRC8B, KIAA0231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8B (T-cell activation leucine repeat-rich protein). | |||||
|
KCT2_MOUSE
|
||||||
| NC score | 0.024798 (rank : 82) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K201, Q5SVC5, Q922L7, Q9CVN1 | Gene names | Kct2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.022842 (rank : 83) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.021975 (rank : 84) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
LRC59_HUMAN
|
||||||
| NC score | 0.021892 (rank : 85) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
LRC32_HUMAN
|
||||||
| NC score | 0.021837 (rank : 86) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14392, Q86V06 | Gene names | LRRC32, GARP | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 32 precursor (GARP protein) (Garpin) (Glycoprotein A repetitions predominant). | |||||
|
LRC59_MOUSE
|
||||||
| NC score | 0.021760 (rank : 87) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q922Q8, Q3TJ35, Q3TLC7, Q3TWT9, Q3TX86 | Gene names | Lrrc59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.020164 (rank : 88) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
TRH_MOUSE
|
||||||
| NC score | 0.019741 (rank : 89) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62361, Q7TMT4 | Gene names | Trh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroliberin precursor [Contains: Prothyroliberin; Thyroliberin (Thyrotropin-releasing hormone) (TRH) (Thyrotropin-releasing factor) (TRF) (TSH-releasing factor) (Protirelin)]. | |||||
|
LGR5_HUMAN
|
||||||
| NC score | 0.018646 (rank : 90) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75473, Q9UP75 | Gene names | LGR5, GPR49, GPR67 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 5 precursor (Orphan G-protein coupled receptor HG38) (G-protein coupled receptor 49) (G-protein coupled receptor 67). | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.018547 (rank : 91) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.018473 (rank : 92) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.018119 (rank : 93) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.017943 (rank : 94) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TLR3_MOUSE
|
||||||
| NC score | 0.017561 (rank : 95) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99MB1, Q91ZM4 | Gene names | Tlr3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 3 precursor (CD283 antigen). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.017054 (rank : 96) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CCD22_MOUSE
|
||||||
| NC score | 0.016254 (rank : 97) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIG7, Q8BYH4 | Gene names | Ccdc22, DXImx40e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 22. | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.016219 (rank : 98) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.015992 (rank : 99) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.015018 (rank : 100) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.014970 (rank : 101) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.014899 (rank : 102) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.014728 (rank : 103) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NPHP4_HUMAN
|
||||||
| NC score | 0.014450 (rank : 104) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75161 | Gene names | NPHP4, KIAA0673 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.013907 (rank : 105) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.012674 (rank : 106) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.012189 (rank : 107) | θ value | 0.279714 (rank : 52) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TMM28_HUMAN
|
||||||
| NC score | 0.011240 (rank : 108) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
MOD5_HUMAN
|
||||||
| NC score | 0.010762 (rank : 109) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |

|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.010132 (rank : 110) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
GA2L1_MOUSE
|
||||||
| NC score | 0.010104 (rank : 111) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.009737 (rank : 112) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.009733 (rank : 113) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.009536 (rank : 114) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.009522 (rank : 115) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.009481 (rank : 116) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.009319 (rank : 117) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.009289 (rank : 118) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.009257 (rank : 119) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GATA2_MOUSE
|
||||||
| NC score | 0.009111 (rank : 120) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
ORC3_HUMAN
|
||||||
| NC score | 0.008789 (rank : 121) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
F100A_HUMAN
|
||||||
| NC score | 0.008705 (rank : 122) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TB05, Q71MF6 | Gene names | FAM100A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
LEG3_HUMAN
|
||||||
| NC score | 0.008669 (rank : 123) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.008206 (rank : 124) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.008198 (rank : 125) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
GATA2_HUMAN
|
||||||
| NC score | 0.007988 (rank : 126) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.007635 (rank : 127) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.007615 (rank : 128) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.007260 (rank : 129) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
EMIL3_HUMAN
|
||||||
| NC score | 0.006654 (rank : 130) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NT22, Q76KT4 | Gene names | EMILIN3, C20orf130, EMILIN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface-located protein 5) (Elastin microfibril interfacer 5). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.006559 (rank : 131) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DIRA3_HUMAN
|
||||||
| NC score | 0.006199 (rank : 132) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.006173 (rank : 133) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.006133 (rank : 134) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.005584 (rank : 135) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.005239 (rank : 136) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.004816 (rank : 137) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.004211 (rank : 138) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.003355 (rank : 139) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.003196 (rank : 140) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.002975 (rank : 141) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.002877 (rank : 142) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
LRRK1_HUMAN
|
||||||
| NC score | 0.002578 (rank : 143) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q38SD2, Q6NVH5, Q6NYC0, Q6ZNL9, Q6ZNM9, Q96JN5, Q9H5S3 | Gene names | LRRK1, KIAA1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.001827 (rank : 144) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.001815 (rank : 145) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
PCD20_HUMAN
|
||||||
| NC score | 0.001067 (rank : 146) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.000546 (rank : 147) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.000457 (rank : 148) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.000053 (rank : 149) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
SP5_MOUSE
|
||||||
| NC score | -0.000703 (rank : 150) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_HUMAN
|
||||||
| NC score | -0.001135 (rank : 151) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
NTR2_MOUSE
|
||||||
| NC score | -0.001558 (rank : 152) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70310 | Gene names | Ntsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotensin receptor type 2 (NT-R-2) (Low-affinity levocabastine- sensitive neurotensin receptor) (NTRL). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | -0.001805 (rank : 153) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
KAISO_MOUSE
|
||||||
| NC score | -0.002277 (rank : 154) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BN78, Q8C226, Q9WTZ7 | Gene names | Zbtb33, Kaiso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
TYK2_HUMAN
|
||||||
| NC score | -0.003037 (rank : 155) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29597, Q6QB10, Q96CH0 | Gene names | TYK2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||
|
GLI3_HUMAN
|
||||||
| NC score | -0.003623 (rank : 156) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
MARK1_MOUSE
|
||||||
| NC score | -0.004038 (rank : 157) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHJ5, Q69ZI7 | Gene names | Mark1, Emk3, Kiaa1477 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK1 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 1) (ELKL motif serine/threonine-protein kinase 3). | |||||