Please be patient as the page loads
|
CEP68_HUMAN
|
||||||
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CEP68_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
CEP68_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.932812 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
AKAP6_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 3) | NC score | 0.327848 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 4) | NC score | 0.093000 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 5) | NC score | 0.045611 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.081638 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.089034 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.097902 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.072614 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.035907 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.071010 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.086641 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.027193 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.071690 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.128932 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.040779 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.047411 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.048350 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.062920 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.052069 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.050940 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.067034 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.049024 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CK035_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.056363 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXW0 | Gene names | C11orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35. | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.055594 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.053695 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.065697 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.052327 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.045691 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.039402 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.025194 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.032387 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.055134 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.020068 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.053564 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.059531 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RTN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.043463 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16799, Q16800, Q16801 | Gene names | RTN1, NSP | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.064900 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.054245 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.032651 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.027747 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.020164 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.057275 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
APOA4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.030277 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |
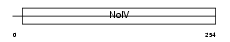
|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.052092 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.042635 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
CD45_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.014781 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.023008 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.025124 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.060610 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.064722 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.035476 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.026338 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.034443 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.034213 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
AAKG2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.021490 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
EPB42_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.016115 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16452 | Gene names | EPB42, E42P | |||
|
Domain Architecture |

|
|||||
| Description | Erythrocyte membrane protein band 4.2 (Erythrocyte protein 4.2) (P4.2). | |||||
|
ERRFI_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.035050 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.059074 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.041953 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.047225 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRTD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.041698 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJW9, Q96CQ2 | Gene names | SERTAD3, RBT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 3 (Replication protein-binding trans- activator) (RPA-binding trans-activator). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.007947 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
APBA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.020289 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.022334 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.012310 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.040295 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.012609 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.056303 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.065731 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SSBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.058261 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P81877, Q8N2Q2, Q9BWW6, Q9Y4T7 | Gene names | SSBP2, SSDP2 | |||
|
Domain Architecture |
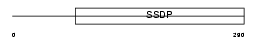
|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
SSBP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.058246 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CYZ8, Q99LX9 | Gene names | Ssbp2, Ssdp2 | |||
|
Domain Architecture |
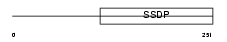
|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.037171 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.033494 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.012898 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.044643 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DCC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.012613 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.012250 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
ILDR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.024655 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CBR1, Q6PFB3, Q8CB39, Q91VS0 | Gene names | Ildr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.008124 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
OSBL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.010981 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
PIWL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.013561 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TC59, Q96SW6, Q9NW28 | Gene names | PIWIL2, HILI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
SDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.035407 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18827, Q96HB7 | Gene names | SDC1, SDC | |||
|
Domain Architecture |
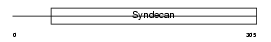
|
|||||
| Description | Syndecan-1 precursor (SYND1) (CD138 antigen). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.043162 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TRI56_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.014068 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
ZN646_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.001622 (rank : 133) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.018092 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.042141 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GP156_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.021303 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
LRFN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.010809 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULH4, Q5SYP9 | Gene names | LRFN2, KIAA1246 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 2 precursor. | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.055058 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.048927 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MYPN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.017907 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
PAG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.023374 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U1F9, Q8BFS1, Q9EQF3 | Gene names | Pag1, Cbp, Pag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (Csk-binding protein) (Transmembrane phosphoprotein Cbp). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.036060 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.024469 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.012343 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.041369 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.024810 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.009551 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
M3K11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.003550 (rank : 131) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.036735 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NUPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.033619 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.030406 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RAVR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.014198 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.046661 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SSH3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.010462 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
TULP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.015392 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
ZDH19_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.010902 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810M5 | Gene names | Zdhhc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.004725 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.023015 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.017628 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.022872 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
LRFN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.009828 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TG9, Q9CYK3 | Gene names | Lrfn2, Kiaa1246 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 2 precursor. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.057522 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MYPN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.018225 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.014761 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.050173 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PA24B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.007065 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PDCD7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.028839 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.016850 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.019447 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.031022 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PTN5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.006902 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
ROR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.001091 (rank : 134) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01973, Q92776 | Gene names | ROR1, NTRKR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.012428 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.014841 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZN219_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.002427 (rank : 132) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052574 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.050317 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
DRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.059303 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.050470 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051981 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.051981 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
CEP68_MOUSE
|
||||||
| NC score | 0.932812 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
AKAP6_HUMAN
|
||||||
| NC score | 0.327848 (rank : 3) | θ value | 1.9326e-10 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.128932 (rank : 4) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.097902 (rank : 5) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.093000 (rank : 6) | θ value | 0.00298849 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.089034 (rank : 7) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.086641 (rank : 8) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.081638 (rank : 9) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.072614 (rank : 10) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.071690 (rank : 11) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.071010 (rank : 12) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.067034 (rank : 13) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.065731 (rank : 14) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.065697 (rank : 15) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.064900 (rank : 16) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.064722 (rank : 17) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.062920 (rank : 18) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.060610 (rank : 19) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.059531 (rank : 20) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.059303 (rank : 21) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.059074 (rank : 22) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SSBP2_HUMAN
|
||||||
| NC score | 0.058261 (rank : 23) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P81877, Q8N2Q2, Q9BWW6, Q9Y4T7 | Gene names | SSBP2, SSDP2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
SSBP2_MOUSE
|
||||||
| NC score | 0.058246 (rank : 24) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CYZ8, Q99LX9 | Gene names | Ssbp2, Ssdp2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 2 (Sequence-specific single- stranded-DNA-binding protein 2). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.057522 (rank : 25) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.057275 (rank : 26) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
CK035_HUMAN
|
||||||
| NC score | 0.056363 (rank : 27) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXW0 | Gene names | C11orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.056303 (rank : 28) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.055594 (rank : 29) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.055134 (rank : 30) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.055058 (rank : 31) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.054245 (rank : 32) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.053695 (rank : 33) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.053564 (rank : 34) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.052574 (rank : 35) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.052327 (rank : 36) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.052092 (rank : 37) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.052069 (rank : 38) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.051981 (rank : 39) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.051981 (rank : 40) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.050940 (rank : 41) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.050470 (rank : 42) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.050317 (rank : 43) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.050173 (rank : 44) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.049024 (rank : 45) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.048927 (rank : 46) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.048350 (rank : 47) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.047411 (rank : 48) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.047225 (rank : 49) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.046661 (rank : 50) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.045691 (rank : 51) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.045611 (rank : 52) | θ value | 0.00298849 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.044643 (rank : 53) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RTN1_HUMAN
|
||||||
| NC score | 0.043463 (rank : 54) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16799, Q16800, Q16801 | Gene names | RTN1, NSP | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.043162 (rank : 55) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.042635 (rank : 56) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.042141 (rank : 57) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.041953 (rank : 58) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SRTD3_HUMAN
|
||||||
| NC score | 0.041698 (rank : 59) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJW9, Q96CQ2 | Gene names | SERTAD3, RBT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 3 (Replication protein-binding trans- activator) (RPA-binding trans-activator). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.041369 (rank : 60) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.040779 (rank : 61) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.040295 (rank : 62) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.039402 (rank : 63) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.037171 (rank : 64) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.036735 (rank : 65) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.036060 (rank : 66) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.035907 (rank : 67) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.035476 (rank : 68) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SDC1_HUMAN
|
||||||
| NC score | 0.035407 (rank : 69) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18827, Q96HB7 | Gene names | SDC1, SDC | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-1 precursor (SYND1) (CD138 antigen). | |||||
|
ERRFI_MOUSE
|
||||||
| NC score | 0.035050 (rank : 70) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.034443 (rank : 71) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.034213 (rank : 72) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
NUPL_HUMAN
|
||||||
| NC score | 0.033619 (rank : 73) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.033494 (rank : 74) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.032651 (rank : 75) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.032387 (rank : 76) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.031022 (rank : 77) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.030406 (rank : 78) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
APOA4_MOUSE
|
||||||
| NC score | 0.030277 (rank : 79) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
PDCD7_HUMAN
|
||||||
| NC score | 0.028839 (rank : 80) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.027747 (rank : 81) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.027193 (rank : 82) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.026338 (rank : 83) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.025194 (rank : 84) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.025124 (rank : 85) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.024810 (rank : 86) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
ILDR1_MOUSE
|
||||||
| NC score | 0.024655 (rank : 87) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CBR1, Q6PFB3, Q8CB39, Q91VS0 | Gene names | Ildr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.024469 (rank : 88) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
PAG1_MOUSE
|
||||||
| NC score | 0.023374 (rank : 89) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U1F9, Q8BFS1, Q9EQF3 | Gene names | Pag1, Cbp, Pag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (Csk-binding protein) (Transmembrane phosphoprotein Cbp). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.023015 (rank : 90) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.023008 (rank : 91) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.022872 (rank : 92) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.022334 (rank : 93) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
AAKG2_HUMAN
|
||||||
| NC score | 0.021490 (rank : 94) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.021303 (rank : 95) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
APBA2_MOUSE
|
||||||
| NC score | 0.020289 (rank : 96) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.020164 (rank : 97) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.020068 (rank : 98) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.019447 (rank : 99) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
MYPN_MOUSE
|
||||||
| NC score | 0.018225 (rank : 100) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.018092 (rank : 101) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
MYPN_HUMAN
|
||||||
| NC score | 0.017907 (rank : 102) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.017628 (rank : 103) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.016850 (rank : 104) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
EPB42_HUMAN
|
||||||
| NC score | 0.016115 (rank : 105) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16452 | Gene names | EPB42, E42P | |||
|
Domain Architecture |

|
|||||
| Description | Erythrocyte membrane protein band 4.2 (Erythrocyte protein 4.2) (P4.2). | |||||
|
TULP1_HUMAN
|
||||||
| NC score | 0.015392 (rank : 106) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.014841 (rank : 107) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.014781 (rank : 108) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.014761 (rank : 109) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
RAVR1_MOUSE
|
||||||
| NC score | 0.014198 (rank : 110) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.014068 (rank : 111) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
PIWL2_HUMAN
|
||||||
| NC score | 0.013561 (rank : 112) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TC59, Q96SW6, Q9NW28 | Gene names | PIWIL2, HILI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.012898 (rank : 113) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.012613 (rank : 114) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.012609 (rank : 115) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.012428 (rank : 116) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.012343 (rank : 117) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.012310 (rank : 118) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.012250 (rank : 119) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
OSBL1_HUMAN
|
||||||
| NC score | 0.010981 (rank : 120) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
ZDH19_MOUSE
|
||||||
| NC score | 0.010902 (rank : 121) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810M5 | Gene names | Zdhhc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
LRFN2_HUMAN
|
||||||
| NC score | 0.010809 (rank : 122) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULH4, Q5SYP9 | Gene names | LRFN2, KIAA1246 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 2 precursor. | |||||
|
SSH3_MOUSE
|
||||||
| NC score | 0.010462 (rank : 123) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
LRFN2_MOUSE
|
||||||
| NC score | 0.009828 (rank : 124) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TG9, Q9CYK3 | Gene names | Lrfn2, Kiaa1246 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 2 precursor. | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.009551 (rank : 125) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.008124 (rank : 126) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.007947 (rank : 127) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
PA24B_HUMAN
|
||||||
| NC score | 0.007065 (rank : 128) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.006902 (rank : 129) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.004725 (rank : 130) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
M3K11_HUMAN
|
||||||
| NC score | 0.003550 (rank : 131) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
ZN219_HUMAN
|
||||||
| NC score | 0.002427 (rank : 132) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
ZN646_HUMAN
|
||||||
| NC score | 0.001622 (rank : 133) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
ROR1_HUMAN
|
||||||
| NC score | 0.001091 (rank : 134) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01973, Q92776 | Gene names | ROR1, NTRKR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1). | |||||