Please be patient as the page loads
|
RGP1_HUMAN
|
||||||
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RGP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.958513 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 3) | NC score | 0.391840 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
LRC31_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 4) | NC score | 0.419317 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 5) | NC score | 0.428046 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 6) | NC score | 0.383626 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
RINI_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 7) | NC score | 0.453711 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
CARD4_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 8) | NC score | 0.431347 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
LRC34_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 9) | NC score | 0.463473 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
RINI_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 10) | NC score | 0.443924 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 11) | NC score | 0.375808 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 12) | NC score | 0.421028 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 13) | NC score | 0.406657 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 14) | NC score | 0.365348 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
LRC34_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 15) | NC score | 0.439758 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 16) | NC score | 0.369171 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 17) | NC score | 0.376079 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 18) | NC score | 0.376609 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 19) | NC score | 0.359226 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 20) | NC score | 0.318099 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 21) | NC score | 0.156837 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CC47_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 22) | NC score | 0.146566 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
TRM6_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 23) | NC score | 0.117053 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJA5, Q76P92, Q9BQV5, Q9ULR7 | Gene names | TRM6, KIAA1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 24) | NC score | 0.119131 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ZN694_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 25) | NC score | 0.005394 (rank : 150) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
TR150_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.060891 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TRM6_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.111731 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CE96, Q3TJZ8, Q3TME7, Q6ZPW8, Q80Y59, Q8CEU0 | Gene names | Trm6, Kiaa1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.105754 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.341634 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.102211 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.140671 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.043810 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.078621 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.095575 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.282103 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.072685 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.091314 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.038200 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.082212 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
AR13B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.024700 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
CCD47_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.132696 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
FXL14_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.057219 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N1E6 | Gene names | FBXL14, FBL14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
FXL14_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.057188 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BID8, Q3U2Q9, Q8R5H7, Q8VDT7, Q922N5 | Gene names | Fbxl14, Fbl14, Ppa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
LGR6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.015574 (rank : 142) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBX8, Q5T509, Q5T512, Q6UY15, Q86VU0, Q96K69, Q9BYD7 | Gene names | LGR6 | |||
|
Domain Architecture |
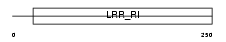
|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 6 (VTS20631). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.023483 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.078428 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.033850 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.019670 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.070562 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.053105 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.028205 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SIRPG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.023095 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P1W8, Q8WWA5, Q9NQK8 | Gene names | SIRPG, SIRPB2 | |||
|
Domain Architecture |
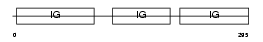
|
|||||
| Description | Signal-regulatory protein gamma precursor (Signal-regulatory protein beta-2) (SIRP-beta-2) (SIRP-b2) (CD172g antigen). | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.021654 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
NUCB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.031949 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.024881 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.020919 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.039642 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.035280 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.019411 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
LETM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.030206 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.027002 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.022562 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.032526 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
41_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.014385 (rank : 145) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.015548 (rank : 143) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CF154_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.300221 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
F10A4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.049466 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
LRRC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.023083 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDB8, Q9CX04 | Gene names | Lrrc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 2. | |||||
|
NASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.052500 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PA24A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.016940 (rank : 136) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.021417 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
SAS10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.047165 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.041764 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.021450 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.023580 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
AL2SA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.036470 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.039030 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.019591 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.035333 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.034496 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
PSMD2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.039727 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.030034 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
TSGA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.021228 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
ZKSC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.003118 (rank : 153) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||
|
ANLN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.036493 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.033145 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.045076 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.034396 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.042572 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.048197 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.045601 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
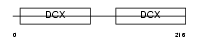
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
DMXL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.017970 (rank : 134) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
FAF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.024493 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.016641 (rank : 139) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.049384 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.005231 (rank : 151) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
RAGE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.009790 (rank : 148) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62151 | Gene names | Ager, Rage | |||
|
Domain Architecture |

|
|||||
| Description | Advanced glycosylation end product-specific receptor precursor (Receptor for advanced glycosylation end products). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.052056 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
ANR38_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.013043 (rank : 146) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.023874 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.019300 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FAF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.023548 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
RNF25_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.016722 (rank : 137) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |
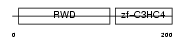
|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
SIRB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.014513 (rank : 144) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00241, Q5TFR0, Q8TB12, Q9H1U5, Q9Y4V0 | Gene names | SIRPB1 | |||
|
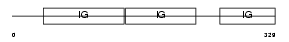
Domain Architecture |
|
|||||
| Description | Signal-regulatory protein beta-1 precursor (SIRP-beta-1) (CD172b antigen). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.016671 (rank : 138) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
TMOD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.047217 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49813, Q9ERR9 | Gene names | Tmod1, Tmod | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-1 (Erythrocyte tropomodulin) (E-Tmod). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.030190 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.063143 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ZBT10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.005219 (rank : 152) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96DT7, Q86W96, Q8IXI9, Q96MH9 | Gene names | ZBTB10, RINZF, RINZFC | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 10 (Zinc finger protein RIN ZF). | |||||
|
ZBT24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | -0.000192 (rank : 154) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.016372 (rank : 140) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CHMP6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.015900 (rank : 141) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.019804 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.068856 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.038042 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.018294 (rank : 133) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NEK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | -0.001766 (rank : 155) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
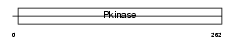
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.024904 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.010374 (rank : 147) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PSA7L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.006997 (rank : 149) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWH6 | Gene names | Psma8, Psma7l | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7-like (EC 3.4.25.1). | |||||
|
PSMD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.024059 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
SMG5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.037112 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.026066 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.017922 (rank : 135) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.051952 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051860 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
CAR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.119307 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.057074 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.053136 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.050379 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057180 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.051455 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051434 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.059935 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
LMD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.055281 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.237737 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.230764 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NAL11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.298587 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.302938 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.260180 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.263275 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.301098 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.085977 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.069766 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.059169 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.104571 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.102649 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.098294 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.091452 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.056808 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.054002 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.061716 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCAL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.053627 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.050994 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.053527 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.958513 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.463473 (rank : 3) | θ value | 9.92553e-07 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.453711 (rank : 4) | θ value | 2.61198e-07 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.443924 (rank : 5) | θ value | 2.88788e-06 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.439758 (rank : 6) | θ value | 4.1701e-05 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.431347 (rank : 7) | θ value | 5.81887e-07 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.428046 (rank : 8) | θ value | 2.61198e-07 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.421028 (rank : 9) | θ value | 1.43324e-05 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.419317 (rank : 10) | θ value | 1.17247e-07 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.406657 (rank : 11) | θ value | 2.44474e-05 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.391840 (rank : 12) | θ value | 1.25267e-09 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.383626 (rank : 13) | θ value | 2.61198e-07 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.376609 (rank : 14) | θ value | 0.000158464 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.376079 (rank : 15) | θ value | 0.000121331 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.375808 (rank : 16) | θ value | 3.77169e-06 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.369171 (rank : 17) | θ value | 5.44631e-05 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.365348 (rank : 18) | θ value | 3.19293e-05 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.359226 (rank : 19) | θ value | 0.000461057 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.341634 (rank : 20) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.318099 (rank : 21) | θ value | 0.00134147 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.302938 (rank : 22) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.301098 (rank : 23) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.300221 (rank : 24) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.298587 (rank : 25) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.282103 (rank : 26) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.263275 (rank : 27) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.260180 (rank : 28) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.237737 (rank : 29) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.230764 (rank : 30) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.156837 (rank : 31) | θ value | 0.00134147 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.146566 (rank : 32) | θ value | 0.00298849 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.140671 (rank : 33) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CCD47_HUMAN
|
||||||
| NC score | 0.132696 (rank : 34) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.119307 (rank : 35) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.119131 (rank : 36) | θ value | 0.0193708 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TRM6_HUMAN
|
||||||
| NC score | 0.117053 (rank : 37) | θ value | 0.0148317 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJA5, Q76P92, Q9BQV5, Q9ULR7 | Gene names | TRM6, KIAA1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
TRM6_MOUSE
|
||||||
| NC score | 0.111731 (rank : 38) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CE96, Q3TJZ8, Q3TME7, Q6ZPW8, Q80Y59, Q8CEU0 | Gene names | Trm6, Kiaa1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.105754 (rank : 39) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.104571 (rank : 40) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.102649 (rank : 41) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.102211 (rank : 42) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.098294 (rank : 43) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.095575 (rank : 44) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.091452 (rank : 45) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.091314 (rank : 46) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.085977 (rank : 47) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.082212 (rank : 48) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.078621 (rank : 49) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.078428 (rank : 50) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.072685 (rank : 51) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.070562 (rank : 52) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.069766 (rank : 53) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.068856 (rank : 54) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.063143 (rank : 55) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.061716 (rank : 56) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.060891 (rank : 57) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.059935 (rank : 58) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.059169 (rank : 59) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
FXL14_HUMAN
|
||||||
| NC score | 0.057219 (rank : 60) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N1E6 | Gene names | FBXL14, FBL14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
FXL14_MOUSE
|
||||||
| NC score | 0.057188 (rank : 61) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BID8, Q3U2Q9, Q8R5H7, Q8VDT7, Q922N5 | Gene names | Fbxl14, Fbl14, Ppa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 14 (F-box and leucine-rich repeat protein 14). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.057180 (rank : 62) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.057074 (rank : 63) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.056808 (rank : 64) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
LMD1_HUMAN
|
||||||
| NC score | 0.055281 (rank : 65) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.054002 (rank : 66) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
TCAL3_MOUSE
|
||||||
| NC score | 0.053627 (rank : 67) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.053527 (rank : 68) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.053136 (rank : 69) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.053105 (rank : 70) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.052500 (rank : 71) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.052056 (rank : 72) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.051952 (rank : 73) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.051860 (rank : 74) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.051455 (rank : 75) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.051434 (rank : 76) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.050994 (rank : 77) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.050379 (rank : 78) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.049466 (rank : 79) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.049384 (rank : 80) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.048197 (rank : 81) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
TMOD1_MOUSE
|
||||||
| NC score | 0.047217 (rank : 82) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49813, Q9ERR9 | Gene names | Tmod1, Tmod | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-1 (Erythrocyte tropomodulin) (E-Tmod). | |||||
|
SAS10_HUMAN
|
||||||
| NC score | 0.047165 (rank : 83) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.045601 (rank : 84) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.045076 (rank : 85) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.043810 (rank : 86) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.042572 (rank : 87) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.041764 (rank : 88) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
PSMD2_MOUSE
|
||||||
| NC score | 0.039727 (rank : 89) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.039642 (rank : 90) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.039030 (rank : 91) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.038200 (rank : 92) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.038042 (rank : 93) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
SMG5_MOUSE
|
||||||
| NC score | 0.037112 (rank : 94) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.036493 (rank : 95) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
AL2SA_HUMAN
|
||||||
| NC score | 0.036470 (rank : 96) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.035333 (rank : 97) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.035280 (rank : 98) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.034496 (rank : 99) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.034396 (rank : 100) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.033850 (rank : 101) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.033145 (rank : 102) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.032526 (rank : 103) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
NUCB1_MOUSE
|
||||||
| NC score | 0.031949 (rank : 104) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
LETM1_HUMAN
|
||||||
| NC score | 0.030206 (rank : 105) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.030190 (rank : 106) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.030034 (rank : 107) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.028205 (rank : 108) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.027002 (rank : 109) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.026066 (rank : 110) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.024904 (rank : 111) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.024881 (rank : 112) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
AR13B_HUMAN
|
||||||
| NC score | 0.024700 (rank : 113) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
FAF1_MOUSE
|
||||||
| NC score | 0.024493 (rank : 114) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
PSMD2_HUMAN
|
||||||
| NC score | 0.024059 (rank : 115) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.023874 (rank : 116) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.023580 (rank : 117) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
FAF1_HUMAN
|
||||||
| NC score | 0.023548 (rank : 118) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.023483 (rank : 119) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SIRPG_HUMAN
|
||||||
| NC score | 0.023095 (rank : 120) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P1W8, Q8WWA5, Q9NQK8 | Gene names | SIRPG, SIRPB2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-regulatory protein gamma precursor (Signal-regulatory protein beta-2) (SIRP-beta-2) (SIRP-b2) (CD172g antigen). | |||||
|
LRRC2_MOUSE
|
||||||
| NC score | 0.023083 (rank : 121) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDB8, Q9CX04 | Gene names | Lrrc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 2. | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.022562 (rank : 122) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.021654 (rank : 123) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.021450 (rank : 124) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.021417 (rank : 125) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
TSGA2_HUMAN
|
||||||
| NC score | 0.021228 (rank : 126) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.020919 (rank : 127) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.019804 (rank : 128) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.019670 (rank : 129) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.019591 (rank : 130) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.019411 (rank : 131) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.019300 (rank : 132) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.018294 (rank : 133) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DMXL2_HUMAN
|
||||||
| NC score | 0.017970 (rank : 134) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.017922 (rank : 135) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
PA24A_MOUSE
|
||||||
| NC score | 0.016940 (rank : 136) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
RNF25_HUMAN
|
||||||
| NC score | 0.016722 (rank : 137) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.016671 (rank : 138) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.016641 (rank : 139) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.016372 (rank : 140) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CHMP6_MOUSE
|
||||||
| NC score | 0.015900 (rank : 141) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
LGR6_HUMAN
|
||||||
| NC score | 0.015574 (rank : 142) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBX8, Q5T509, Q5T512, Q6UY15, Q86VU0, Q96K69, Q9BYD7 | Gene names | LGR6 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 6 (VTS20631). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.015548 (rank : 143) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
SIRB1_HUMAN
|
||||||
| NC score | 0.014513 (rank : 144) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00241, Q5TFR0, Q8TB12, Q9H1U5, Q9Y4V0 | Gene names | SIRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-regulatory protein beta-1 precursor (SIRP-beta-1) (CD172b antigen). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.014385 (rank : 145) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
ANR38_MOUSE
|
||||||
| NC score | 0.013043 (rank : 146) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.010374 (rank : 147) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
RAGE_MOUSE
|
||||||
| NC score | 0.009790 (rank : 148) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62151 | Gene names | Ager, Rage | |||
|
Domain Architecture |

|
|||||
| Description | Advanced glycosylation end product-specific receptor precursor (Receptor for advanced glycosylation end products). | |||||
|
PSA7L_MOUSE
|
||||||
| NC score | 0.006997 (rank : 149) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWH6 | Gene names | Psma8, Psma7l | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7-like (EC 3.4.25.1). | |||||
|
ZN694_HUMAN
|
||||||
| NC score | 0.005394 (rank : 150) | θ value | 0.0330416 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.005231 (rank : 151) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
ZBT10_HUMAN
|
||||||
| NC score | 0.005219 (rank : 152) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96DT7, Q86W96, Q8IXI9, Q96MH9 | Gene names | ZBTB10, RINZF, RINZFC | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 10 (Zinc finger protein RIN ZF). | |||||
|
ZKSC1_HUMAN
|
||||||
| NC score | 0.003118 (rank : 153) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||
|
ZBT24_HUMAN
|
||||||
| NC score | -0.000192 (rank : 154) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
NEK1_HUMAN
|
||||||
| NC score | -0.001766 (rank : 155) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||