Please be patient as the page loads
|
GA2L1_MOUSE
|
||||||
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GA2L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.953169 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
GA2L1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 3) | NC score | 0.817501 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 4) | NC score | 0.876083 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
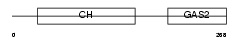
GAS2_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 5) | NC score | 0.832105 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43903, Q6ICV8 | Gene names | GAS2 | |||
|
Domain Architecture |
|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
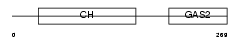
GAS2_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 6) | NC score | 0.832377 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11862 | Gene names | Gas2, Gas-2 | |||
|
Domain Architecture |
|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 7) | NC score | 0.772752 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 8) | NC score | 0.248424 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 9) | NC score | 0.243034 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 10) | NC score | 0.267491 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 11) | NC score | 0.256536 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 12) | NC score | 0.224243 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 13) | NC score | 0.126456 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TSNA1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 14) | NC score | 0.091984 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 15) | NC score | 0.017049 (rank : 172) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 16) | NC score | 0.058064 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 17) | NC score | 0.092589 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.027981 (rank : 157) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.067071 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.050800 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.118830 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.099824 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.036045 (rank : 140) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.103633 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.090377 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.020715 (rank : 169) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.015134 (rank : 176) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.093185 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.061797 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.030559 (rank : 151) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.078850 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
IQGA1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.073768 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
IQGA1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.076436 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.043561 (rank : 131) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.052621 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.025735 (rank : 160) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.104982 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.034524 (rank : 144) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.080053 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.064798 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.033844 (rank : 146) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.053364 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.023765 (rank : 162) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.058031 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.039971 (rank : 135) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.028314 (rank : 155) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.038786 (rank : 136) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
RUSC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.028182 (rank : 156) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.016210 (rank : 173) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
MARK2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.010173 (rank : 187) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.057790 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.032830 (rank : 149) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.010892 (rank : 184) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.035469 (rank : 142) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.035081 (rank : 143) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
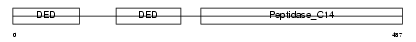
CASP8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.016129 (rank : 174) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CO9A2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.030512 (rank : 152) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.018303 (rank : 171) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
TAU_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.050525 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.073267 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
CO5A1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.036618 (rank : 138) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.052350 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
DLX5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.004420 (rank : 196) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.033932 (rank : 145) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.014873 (rank : 177) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.044389 (rank : 130) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CA147_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.047400 (rank : 128) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MC9, Q5JTS5 | Gene names | C1orf147 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf147. | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.026935 (rank : 158) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.026788 (rank : 159) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.041361 (rank : 133) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
CT128_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.021197 (rank : 168) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
CTG1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.036083 (rank : 139) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78358, Q7LBY4 | Gene names | CTAG1B, CTAG, CTAG1, ESO1, LAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen 1B (L antigen family member 2) (LAGE-2 protein) (Autoimmunogenic cancer/testis antigen NY-ESO-1). | |||||
|
DEN2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.010712 (rank : 185) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ICB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.024504 (rank : 161) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.022311 (rank : 167) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.022699 (rank : 165) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.010104 (rank : 188) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
RARA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.008809 (rank : 191) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.059601 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.084143 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.033455 (rank : 148) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.020062 (rank : 170) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.040590 (rank : 134) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ABL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.003871 (rank : 197) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
ARRD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.013193 (rank : 179) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.023689 (rank : 163) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CNN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.042435 (rank : 132) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.035975 (rank : 141) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DLG7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.016079 (rank : 175) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
ES8L3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.013770 (rank : 178) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.022725 (rank : 164) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
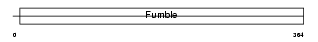
PANK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.008707 (rank : 192) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.032583 (rank : 150) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.013107 (rank : 180) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
S45A3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.012014 (rank : 182) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K0H7, Q8K252, Q8R1I0 | Gene names | Slc45a3, Pcanap6, Prst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 45 member 3 (Prostate cancer-associated protein 6) (Prostein). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.056952 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | -0.000751 (rank : 198) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.008252 (rank : 193) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
BAG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.055673 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.009886 (rank : 189) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.012795 (rank : 181) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.033774 (rank : 147) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CTAG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.028769 (rank : 154) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75638, O75637, Q9BU80, Q9UJ89, Q9Y479 | Gene names | CTAG2, ESO2, LAGE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen 2 (L antigen family member 1) (LAGE-1 protein) (ESO-2 protein). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.029092 (rank : 153) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.005370 (rank : 195) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.075909 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.022539 (rank : 166) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.044957 (rank : 129) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.011885 (rank : 183) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.038521 (rank : 137) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
M3K9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.005561 (rank : 194) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P80192, Q6EH31, Q9H2N5 | Gene names | MAP3K9, MLK1, PRKE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 9 (EC 2.7.11.25) (Mixed lineage kinase 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.073710 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PP4R1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.009783 (rank : 190) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.010245 (rank : 186) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ACTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.062179 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.062638 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.060320 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.061941 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.060672 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.063438 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.061523 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.062065 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
AKAP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.065973 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.050090 (rank : 127) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.051342 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.073561 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.055621 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.060880 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.104658 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.056547 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.052176 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.050461 (rank : 123) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052838 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.065034 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CLMN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.068147 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.060663 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.058099 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DESP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.079046 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DMD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.077875 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.079404 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.066074 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
EVPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.067504 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.071405 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.053068 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.055683 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.061084 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HMGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.052008 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17096, P10910, Q9UKB0 | Gene names | HMGA1, HMGIY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1) (High mobility group protein-R). | |||||
|
IQGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.053285 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.075321 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.076245 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.068432 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
MRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.056092 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050460 (rank : 124) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.050290 (rank : 126) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.064876 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.054746 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.066888 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.070563 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.074559 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.094629 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.052453 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.089962 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.094798 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.073648 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.075446 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.085210 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.078919 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.059254 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.074470 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SFR17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.053873 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.087625 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.074393 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.080623 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.080603 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.073078 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.074344 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.071234 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.069922 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.078149 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.077057 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.077201 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.100941 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.107577 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.103499 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.059333 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.052860 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.080900 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.070605 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.051919 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.064456 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.051415 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.050639 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
UTRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.082207 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.073472 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.070934 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.050990 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.053769 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 198) | NC score | 0.050345 (rank : 125) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
GA2L1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.953169 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.876083 (rank : 3) | θ value | 5.17823e-64 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
GAS2_MOUSE
|
||||||
| NC score | 0.832377 (rank : 4) | θ value | 6.82597e-32 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11862 | Gene names | Gas2, Gas-2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
GAS2_HUMAN
|
||||||
| NC score | 0.832105 (rank : 5) | θ value | 8.06329e-33 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43903, Q6ICV8 | Gene names | GAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.817501 (rank : 6) | θ value | 1.66191e-70 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.772752 (rank : 7) | θ value | 1.28732e-30 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.267491 (rank : 8) | θ value | 4.59992e-12 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.256536 (rank : 9) | θ value | 6.00763e-12 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.248424 (rank : 10) | θ value | 4.91457e-14 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.243034 (rank : 11) | θ value | 4.59992e-12 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.224243 (rank : 12) | θ value | 2.28291e-11 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.126456 (rank : 13) | θ value | 3.19293e-05 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.118830 (rank : 14) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.107577 (rank : 15) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.104982 (rank : 16) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.104658 (rank : 17) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.103633 (rank : 18) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.103499 (rank : 19) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.100941 (rank : 20) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.099824 (rank : 21) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.094798 (rank : 22) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.094629 (rank : 23) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.093185 (rank : 24) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.092589 (rank : 25) | θ value | 0.00665767 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TSNA1_HUMAN
|
||||||
| NC score | 0.091984 (rank : 26) | θ value | 0.00175202 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.090377 (rank : 27) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.089962 (rank : 28) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.087625 (rank : 29) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.085210 (rank : 30) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.084143 (rank : 31) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.082207 (rank : 32) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.080900 (rank : 33) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.080623 (rank : 34) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.080603 (rank : 35) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.080053 (rank : 36) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.079404 (rank : 37) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.079046 (rank : 38) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.078919 (rank : 39) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.078850 (rank : 40) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.078149 (rank : 41) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.077875 (rank : 42) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.077201 (rank : 43) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.077057 (rank : 44) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
IQGA1_MOUSE
|
||||||
| NC score | 0.076436 (rank : 45) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.076245 (rank : 46) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.075909 (rank : 47) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.075446 (rank : 48) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.075321 (rank : 49) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.074559 (rank : 50) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.074470 (rank : 51) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.074393 (rank : 52) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.074344 (rank : 53) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
IQGA1_HUMAN
|
||||||
| NC score | 0.073768 (rank : 54) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.073710 (rank : 55) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.073648 (rank : 56) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.073561 (rank : 57) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.073472 (rank : 58) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.073267 (rank : 59) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.073078 (rank : 60) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.071405 (rank : 61) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.071234 (rank : 62) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.070934 (rank : 63) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.070605 (rank : 64) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.070563 (rank : 65) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.069922 (rank : 66) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.068432 (rank : 67) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.068147 (rank : 68) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.067504 (rank : 69) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.067071 (rank : 70) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.066888 (rank : 71) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.066074 (rank : 72) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
AKAP6_HUMAN
|
||||||
| NC score | 0.065973 (rank : 73) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.065034 (rank : 74) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.064876 (rank : 75) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.064798 (rank : 76) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.064456 (rank : 77) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ACTN3_MOUSE
|
||||||
| NC score | 0.063438 (rank : 78) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| NC score | 0.062638 (rank : 79) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_HUMAN
|
||||||
| NC score | 0.062179 (rank : 80) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| NC score | 0.062065 (rank : 81) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.061941 (rank : 82) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.061797 (rank : 83) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ACTN4_HUMAN
|
||||||
| NC score | 0.061523 (rank : 84) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.061084 (rank : 85) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.060880 (rank : 86) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
ACTN3_HUMAN
|
||||||
| NC score | 0.060672 (rank : 87) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.060663 (rank : 88) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.060320 (rank : 89) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.059601 (rank : 90) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.059333 (rank : 91) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.059254 (rank : 92) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.058099 (rank : 93) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.058064 (rank : 94) | θ value | 0.00390308 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.058031 (rank : 95) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.057790 (rank : 96) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.056952 (rank : 97) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.056547 (rank : 98) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.056092 (rank : 99) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.055683 (rank : 100) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.055673 (rank : 101) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.055621 (rank : 102) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.054746 (rank : 103) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.053873 (rank : 104) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.053769 (rank : 105) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.053364 (rank : 106) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
IQGA2_HUMAN
|
||||||
| NC score | 0.053285 (rank : 107) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.053068 (rank : 108) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.052860 (rank : 109) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.052838 (rank : 110) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.052621 (rank : 111) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.052453 (rank : 112) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.052350 (rank : 113) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.052176 (rank : 114) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
HMGA1_HUMAN
|
||||||
| NC score | 0.052008 (rank : 115) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17096, P10910, Q9UKB0 | Gene names | HMGA1, HMGIY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1) (High mobility group protein-R). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.051919 (rank : 116) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.051415 (rank : 117) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.051342 (rank : 118) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.050990 (rank : 119) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.050800 (rank : 120) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.050639 (rank : 121) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.050525 (rank : 122) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.050461 (rank : 123) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.050460 (rank : 124) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.050345 (rank : 125) | θ value | θ > 10 (rank : 198) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.050290 (rank : 126) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.050090 (rank : 127) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CA147_HUMAN
|
||||||
| NC score | 0.047400 (rank : 128) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MC9, Q5JTS5 | Gene names | C1orf147 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf147. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.044957 (rank : 129) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.044389 (rank : 130) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.043561 (rank : 131) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CNN3_MOUSE
|
||||||
| NC score | 0.042435 (rank : 132) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.041361 (rank : 133) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.040590 (rank : 134) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.039971 (rank : 135) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.038786 (rank : 136) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.038521 (rank : 137) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CO5A1_MOUSE
|
||||||
| NC score | 0.036618 (rank : 138) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CTG1B_HUMAN
|
||||||
| NC score | 0.036083 (rank : 139) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78358, Q7LBY4 | Gene names | CTAG1B, CTAG, CTAG1, ESO1, LAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen 1B (L antigen family member 2) (LAGE-2 protein) (Autoimmunogenic cancer/testis antigen NY-ESO-1). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.036045 (rank : 140) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.035975 (rank : 141) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.035469 (rank : 142) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.035081 (rank : 143) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.034524 (rank : 144) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.033932 (rank : 145) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.033844 (rank : 146) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.033774 (rank : 147) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.033455 (rank : 148) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.032830 (rank : 149) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.032583 (rank : 150) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.030559 (rank : 151) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CO9A2_HUMAN
|
||||||
| NC score | 0.030512 (rank : 152) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.029092 (rank : 153) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CTAG2_HUMAN
|
||||||
| NC score | 0.028769 (rank : 154) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75638, O75637, Q9BU80, Q9UJ89, Q9Y479 | Gene names | CTAG2, ESO2, LAGE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen 2 (L antigen family member 1) (LAGE-1 protein) (ESO-2 protein). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.028314 (rank : 155) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
RUSC1_HUMAN
|
||||||
| NC score | 0.028182 (rank : 156) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.027981 (rank : 157) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.026935 (rank : 158) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.026788 (rank : 159) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.025735 (rank : 160) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ICB1_MOUSE
|
||||||
| NC score | 0.024504 (rank : 161) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.023765 (rank : 162) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.023689 (rank : 163) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.022725 (rank : 164) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.022699 (rank : 165) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.022539 (rank : 166) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.022311 (rank : 167) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
CT128_HUMAN
|
||||||
| NC score | 0.021197 (rank : 168) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.020715 (rank : 169) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.020062 (rank : 170) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.018303 (rank : 171) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.017049 (rank : 172) | θ value | 0.00298849 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.016210 (rank : 173) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.016129 (rank : 174) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
DLG7_MOUSE
|
||||||
| NC score | 0.016079 (rank : 175) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.015134 (rank : 176) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.014873 (rank : 177) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
ES8L3_HUMAN
|
||||||
| NC score | 0.013770 (rank : 178) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.013193 (rank : 179) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.013107 (rank : 180) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.012795 (rank : 181) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
S45A3_MOUSE
|
||||||
| NC score | 0.012014 (rank : 182) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K0H7, Q8K252, Q8R1I0 | Gene names | Slc45a3, Pcanap6, Prst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 45 member 3 (Prostate cancer-associated protein 6) (Prostein). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.011885 (rank : 183) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.010892 (rank : 184) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
DEN2A_HUMAN
|
||||||
| NC score | 0.010712 (rank : 185) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.010245 (rank : 186) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
MARK2_MOUSE
|
||||||
| NC score | 0.010173 (rank : 187) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.010104 (rank : 188) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.009886 (rank : 189) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
PP4R1_MOUSE
|
||||||
| NC score | 0.009783 (rank : 190) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
RARA_HUMAN
|
||||||
| NC score | 0.008809 (rank : 191) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
PANK1_MOUSE
|
||||||
| NC score | 0.008707 (rank : 192) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.008252 (rank : 193) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
M3K9_HUMAN
|
||||||
| NC score | 0.005561 (rank : 194) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P80192, Q6EH31, Q9H2N5 | Gene names | MAP3K9, MLK1, PRKE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 9 (EC 2.7.11.25) (Mixed lineage kinase 1). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.005370 (rank : 195) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DLX5_MOUSE
|
||||||
| NC score | 0.004420 (rank : 196) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
ABL2_HUMAN
|
||||||
| NC score | 0.003871 (rank : 197) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | -0.000751 (rank : 198) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||