Please be patient as the page loads
|
ARRD1_HUMAN
|
||||||
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARRD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
ARRD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.973047 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99KN1, Q3TDT6, Q8VEG7 | Gene names | Arrdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
ARRD2_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 3) | NC score | 0.752141 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D668, Q3UVK8, Q3UXC6 | Gene names | Arrdc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 2. | |||||
|
ARRD2_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 4) | NC score | 0.743263 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBH0, O95895, Q6ZRV9, Q8WYG6 | Gene names | ARRDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 2. | |||||
|
ARRD4_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 5) | NC score | 0.733857 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NCT1, Q6NSI9 | Gene names | ARRDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 4. | |||||
|
ARRD3_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 6) | NC score | 0.724447 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TPQ9, Q4KMN0, Q80TE6, Q8K0L7 | Gene names | Arrdc3, Kiaa1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
ARRD3_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 7) | NC score | 0.721563 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96B67, Q9P2H1 | Gene names | ARRDC3, KIAA1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
TXNIP_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 8) | NC score | 0.696128 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3M7, Q16226, Q6PML0, Q9BXG9 | Gene names | TXNIP, VDUP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin-interacting protein (Vitamin D3 up-regulated protein 1) (Thioredoxin-binding protein 2). | |||||
|
TXNIP_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 9) | NC score | 0.693026 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BG60, Q8BGQ0, Q8K2B2, Q9DC00, Q9EP90, Q9R115 | Gene names | Txnip, Vdup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin-interacting protein (Vitamin D3 up-regulated protein 1). | |||||
|
ARRD5_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 10) | NC score | 0.523572 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q497K5 | Gene names | Arrdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 5. | |||||
|
ARRB2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.117430 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32121, Q0Z8D3, Q2PP19, Q6ICT3, Q8N7Y2, Q9UEQ6 | Gene names | ARRB2, ARB2, ARR2 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-arrestin-2 (Arrestin beta 2). | |||||
|
ARRB2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.107866 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91YI4, Q3TCM2, Q5F2D8, Q5F2E0 | Gene names | Arrb2 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-arrestin-2 (Arrestin beta 2). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.038225 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ARRB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.115608 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49407, O75625, O75630, Q2PP20, Q9BTK8 | Gene names | ARRB1, ARR1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-arrestin-1 (Arrestin beta 1). | |||||
|
DSCR3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.138337 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35075, Q542V5 | Gene names | Dscr3, Dcra, Dscra | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome critical region protein 3 homolog (Down syndrome critical region protein A homolog). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.045382 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DSCR3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.135726 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14972 | Gene names | DSCR3, DCRA, DSCRA | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome critical region protein 3 (Down syndrome critical region protein A). | |||||
|
ARRB1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.101323 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BWG8, Q3UH95, Q8BTJ5 | Gene names | Arrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-arrestin-1 (Arrestin beta 1). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.033562 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.049252 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
CA106_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.048459 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.033381 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.038906 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.030443 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
PEF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.022774 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PAX3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.014350 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.039716 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.039169 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.003805 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.045382 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PASK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.003679 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.013212 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
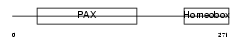
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.030349 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PRGC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.047550 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.011596 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
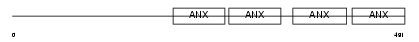
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
CC062_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.032320 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.021819 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.001289 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.021805 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FYB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.027474 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.024229 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CA106_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.039215 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.019216 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PRGC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.046481 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.031034 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.035936 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
BMP3B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.017743 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97737 | Gene names | Gdf10, Bmp3b | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.022427 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.010983 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.020948 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.027428 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
ADAM8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.004382 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
BMP3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.017044 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55107, Q9UCX6 | Gene names | GDF10, BMP3B | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10) (Bone-inducing protein) (BIP). | |||||
|
BMP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.015048 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12645 | Gene names | BMP3 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3 precursor (BMP-3) (Osteogenin) (BMP-3A). | |||||
|
BRSK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.000512 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.010199 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
LBXCO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.015251 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.011227 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.009729 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PTCD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.018914 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75127, Q66K60, Q9UDV2 | Gene names | PTCD1, KIAA0632 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pentatricopeptide repeat protein 1. | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.030316 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.024991 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.007649 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.015292 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
CEND_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.024997 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N111, Q9NYM6 | Gene names | CEND1, BM88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
GA2L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.013193 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.017261 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.014081 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
AB1IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.009301 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
CN004_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.017625 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.016422 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
LIMD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.005244 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
LRC50_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.011509 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.022037 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.031602 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
ARRC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.062368 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36575, Q5JT23, Q5JT24, Q6IBF5, Q96EN2 | Gene names | ARR3, ARRX, CAR | |||
|
Domain Architecture |
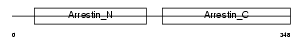
|
|||||
| Description | Arrestin-C (Cone arrestin) (cArr) (Retinal cone arrestin-3) (X- arrestin) (C-arrestin). | |||||
|
ARRC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.086424 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQP6 | Gene names | Arr3 | |||
|
Domain Architecture |
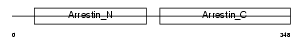
|
|||||
| Description | Arrestin-C (Cone arrestin) (cArr) (Retinal cone arrestin-3). | |||||
|
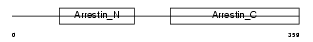
ARRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.061924 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |
|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
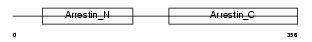
ARRS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.064800 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20443, Q91W62 | Gene names | Sag | |||
|
Domain Architecture |
|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
ARRD1_MOUSE
|
||||||
| NC score | 0.973047 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99KN1, Q3TDT6, Q8VEG7 | Gene names | Arrdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
ARRD2_MOUSE
|
||||||
| NC score | 0.752141 (rank : 3) | θ value | 2.87452e-22 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D668, Q3UVK8, Q3UXC6 | Gene names | Arrdc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 2. | |||||
|
ARRD2_HUMAN
|
||||||
| NC score | 0.743263 (rank : 4) | θ value | 1.86321e-21 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBH0, O95895, Q6ZRV9, Q8WYG6 | Gene names | ARRDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 2. | |||||
|
ARRD4_HUMAN
|
||||||
| NC score | 0.733857 (rank : 5) | θ value | 1.2077e-20 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NCT1, Q6NSI9 | Gene names | ARRDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 4. | |||||
|
ARRD3_MOUSE
|
||||||
| NC score | 0.724447 (rank : 6) | θ value | 3.51386e-20 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TPQ9, Q4KMN0, Q80TE6, Q8K0L7 | Gene names | Arrdc3, Kiaa1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
ARRD3_HUMAN
|
||||||
| NC score | 0.721563 (rank : 7) | θ value | 4.58923e-20 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96B67, Q9P2H1 | Gene names | ARRDC3, KIAA1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
TXNIP_HUMAN
|
||||||
| NC score | 0.696128 (rank : 8) | θ value | 2.60593e-15 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3M7, Q16226, Q6PML0, Q9BXG9 | Gene names | TXNIP, VDUP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin-interacting protein (Vitamin D3 up-regulated protein 1) (Thioredoxin-binding protein 2). | |||||
|
TXNIP_MOUSE
|
||||||
| NC score | 0.693026 (rank : 9) | θ value | 1.29331e-14 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BG60, Q8BGQ0, Q8K2B2, Q9DC00, Q9EP90, Q9R115 | Gene names | Txnip, Vdup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin-interacting protein (Vitamin D3 up-regulated protein 1). | |||||
|
ARRD5_MOUSE
|
||||||
| NC score | 0.523572 (rank : 10) | θ value | 7.59969e-07 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q497K5 | Gene names | Arrdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 5. | |||||
|
DSCR3_MOUSE
|
||||||
| NC score | 0.138337 (rank : 11) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35075, Q542V5 | Gene names | Dscr3, Dcra, Dscra | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome critical region protein 3 homolog (Down syndrome critical region protein A homolog). | |||||
|
DSCR3_HUMAN
|
||||||
| NC score | 0.135726 (rank : 12) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14972 | Gene names | DSCR3, DCRA, DSCRA | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome critical region protein 3 (Down syndrome critical region protein A). | |||||
|
ARRB2_HUMAN
|
||||||
| NC score | 0.117430 (rank : 13) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32121, Q0Z8D3, Q2PP19, Q6ICT3, Q8N7Y2, Q9UEQ6 | Gene names | ARRB2, ARB2, ARR2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-arrestin-2 (Arrestin beta 2). | |||||
|
ARRB1_HUMAN
|
||||||
| NC score | 0.115608 (rank : 14) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49407, O75625, O75630, Q2PP20, Q9BTK8 | Gene names | ARRB1, ARR1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-arrestin-1 (Arrestin beta 1). | |||||
|
ARRB2_MOUSE
|
||||||
| NC score | 0.107866 (rank : 15) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91YI4, Q3TCM2, Q5F2D8, Q5F2E0 | Gene names | Arrb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-arrestin-2 (Arrestin beta 2). | |||||
|
ARRB1_MOUSE
|
||||||
| NC score | 0.101323 (rank : 16) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BWG8, Q3UH95, Q8BTJ5 | Gene names | Arrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-arrestin-1 (Arrestin beta 1). | |||||
|
ARRC_MOUSE
|
||||||
| NC score | 0.086424 (rank : 17) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQP6 | Gene names | Arr3 | |||
|
Domain Architecture |

|
|||||
| Description | Arrestin-C (Cone arrestin) (cArr) (Retinal cone arrestin-3). | |||||
|
ARRS_MOUSE
|
||||||
| NC score | 0.064800 (rank : 18) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20443, Q91W62 | Gene names | Sag | |||
|
Domain Architecture |

|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
ARRC_HUMAN
|
||||||
| NC score | 0.062368 (rank : 19) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36575, Q5JT23, Q5JT24, Q6IBF5, Q96EN2 | Gene names | ARR3, ARRX, CAR | |||
|
Domain Architecture |

|
|||||
| Description | Arrestin-C (Cone arrestin) (cArr) (Retinal cone arrestin-3) (X- arrestin) (C-arrestin). | |||||
|
ARRS_HUMAN
|
||||||
| NC score | 0.061924 (rank : 20) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |

|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.049252 (rank : 21) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.048459 (rank : 22) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
PRGC1_HUMAN
|
||||||
| NC score | 0.047550 (rank : 23) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
PRGC1_MOUSE
|
||||||
| NC score | 0.046481 (rank : 24) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.045382 (rank : 25) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.045382 (rank : 26) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.039716 (rank : 27) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.039215 (rank : 28) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.039169 (rank : 29) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.038906 (rank : 30) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.038225 (rank : 31) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.035936 (rank : 32) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.033562 (rank : 33) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.033381 (rank : 34) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CC062_HUMAN
|
||||||
| NC score | 0.032320 (rank : 35) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.031602 (rank : 36) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.031034 (rank : 37) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.030443 (rank : 38) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.030349 (rank : 39) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.030316 (rank : 40) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.027474 (rank : 41) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.027428 (rank : 42) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
CEND_HUMAN
|
||||||
| NC score | 0.024997 (rank : 43) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N111, Q9NYM6 | Gene names | CEND1, BM88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.024991 (rank : 44) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.024229 (rank : 45) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
PEF1_HUMAN
|
||||||
| NC score | 0.022774 (rank : 46) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.022427 (rank : 47) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.022037 (rank : 48) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.021819 (rank : 49) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.021805 (rank : 50) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.020948 (rank : 51) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.019216 (rank : 52) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PTCD1_HUMAN
|
||||||
| NC score | 0.018914 (rank : 53) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75127, Q66K60, Q9UDV2 | Gene names | PTCD1, KIAA0632 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pentatricopeptide repeat protein 1. | |||||
|
BMP3B_MOUSE
|
||||||
| NC score | 0.017743 (rank : 54) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97737 | Gene names | Gdf10, Bmp3b | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10). | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.017625 (rank : 55) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.017261 (rank : 56) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
BMP3B_HUMAN
|
||||||
| NC score | 0.017044 (rank : 57) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55107, Q9UCX6 | Gene names | GDF10, BMP3B | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10) (Bone-inducing protein) (BIP). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.016422 (rank : 58) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.015292 (rank : 59) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
LBXCO_HUMAN
|
||||||
| NC score | 0.015251 (rank : 60) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
BMP3_HUMAN
|
||||||
| NC score | 0.015048 (rank : 61) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12645 | Gene names | BMP3 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3 precursor (BMP-3) (Osteogenin) (BMP-3A). | |||||
|
PAX3_HUMAN
|
||||||
| NC score | 0.014350 (rank : 62) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.014081 (rank : 63) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
PAX3_MOUSE
|
||||||
| NC score | 0.013212 (rank : 64) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
GA2L1_MOUSE
|
||||||
| NC score | 0.013193 (rank : 65) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.011596 (rank : 66) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
LRC50_HUMAN
|
||||||
| NC score | 0.011509 (rank : 67) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.011227 (rank : 68) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.010983 (rank : 69) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.010199 (rank : 70) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.009729 (rank : 71) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
AB1IP_MOUSE
|
||||||
| NC score | 0.009301 (rank : 72) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.007649 (rank : 73) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
LIMD1_HUMAN
|
||||||
| NC score | 0.005244 (rank : 74) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
ADAM8_MOUSE
|
||||||
| NC score | 0.004382 (rank : 75) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.003805 (rank : 76) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
PASK_HUMAN
|
||||||
| NC score | 0.003679 (rank : 77) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.001289 (rank : 78) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
BRSK1_HUMAN
|
||||||
| NC score | 0.000512 (rank : 79) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||