Please be patient as the page loads
|
CA106_HUMAN
|
||||||
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CA106_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.965510 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 3) | NC score | 0.479261 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 4) | NC score | 0.478481 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 5) | NC score | 0.438201 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 6) | NC score | 0.431890 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 7) | NC score | 0.091220 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
THSD1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.116539 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.063133 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.059603 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.061964 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.071939 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.061109 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.067770 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.030564 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.067655 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.077638 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
TAU_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.052653 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.018749 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.068466 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.054878 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.020959 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.034647 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.039453 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
FOXGA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.014892 (rank : 139) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55316 | Gene names | FOXG1A, FKHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1A (Forkhead-related protein FKHL2) (Transcription factor BF-2) (Brain factor 2) (BF2) (HFK2). | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.071484 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
MUC15_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.047516 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
CF152_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.031980 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
STK36_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.005708 (rank : 166) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.018138 (rank : 133) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.019427 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CU056_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.040596 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.021945 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
THSD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.061227 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
ATS12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.012760 (rank : 146) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.068741 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.026886 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.069920 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.034673 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MLH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.025942 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
SAKS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.029529 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.020727 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.023506 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.033249 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
N4BP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.025475 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15049, Q7Z6I3 | Gene names | N4BP3, KIAA0341 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.022254 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.009375 (rank : 154) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.011574 (rank : 149) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.026099 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
ARRD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.039215 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.014500 (rank : 141) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.062780 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.039183 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
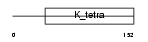
HIC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.002500 (rank : 170) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.035446 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PABP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.031168 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86U42, O43484 | Gene names | PABPN1, PAB2, PABP2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
PABP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.031530 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CCS6, O35935 | Gene names | Pabpn1, Pab2, Pabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.037891 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.022668 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.030961 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
WDR67_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.025723 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NXY1, Q3TBW4, Q3U4F6, Q5KSA3, Q810J7, Q8C3Y2 | Gene names | Wdr67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67 (Protein 4-B-3). | |||||
|
CCD24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.035376 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N4L8, Q6RWT2 | Gene names | CCDC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 24. | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.043158 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.012612 (rank : 147) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.033050 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.048468 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.002426 (rank : 171) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
KSYK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.004497 (rank : 167) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.030870 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MAML1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.021081 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
NAB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.022293 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.028116 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.017933 (rank : 134) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.017914 (rank : 135) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
ENK10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.037434 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10267, Q14273 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Env polyprotein (Envelope polyprotein) (HERV-K10 envelope protein) (HERV-K107 envelope protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.037431 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61566 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Env polyprotein (Envelope polyprotein) (HERV-K101 envelope protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.037393 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61567 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Env polyprotein (Envelope polyprotein) (HERV-K102 envelope protein) (HERV-K(III) envelope protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.034742 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61570 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.035055 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61565 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (EnvK1 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.034888 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q69384, Q8TD93, Q9UBU4, Q9YNA5, Q9YNA9 | Gene names | ERVK6 | |||
|
Domain Architecture |

|
|||||
| Description | HERV-K_7p22.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K(HML-2.HOM) envelope protein) (HERV-K108 envelope protein) (HERV-K(C7) envelope protein) (EnvK2 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.034849 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O71037, Q69386, Q9YNA7 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_19q12 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K(C19) envelope protein) (EnvK3 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.034921 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKH3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_6q14.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K109 envelope protein) (HERV-K(C6) envelope protein) (EnvK4 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.034859 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q902F8 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_8p23.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K115 envelope protein) (EnvK6 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.017666 (rank : 136) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.050168 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.035894 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.003429 (rank : 169) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
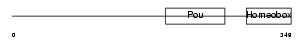
PO2F2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.008422 (rank : 157) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
POK12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.028360 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63135 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pol protein (HERV-K102 Pol protein) (HERV-K(III) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
PP4R1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.036775 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.007906 (rank : 160) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SP100_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.019005 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35892, O35897, O88392, O88395 | Gene names | Sp100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.019263 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.015810 (rank : 137) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ZN250_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | -0.001109 (rank : 172) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNU6, Q8BJ57 | Gene names | Znf250, Zfp647, Znf647 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 250 (Zinc finger protein 647). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.013646 (rank : 145) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
BCN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.015754 (rank : 138) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88597 | Gene names | Becn1 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.044701 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.041834 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DTNB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.025399 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.008607 (rank : 156) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.030970 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.023354 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PRGR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.006114 (rank : 163) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
RAVR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.011028 (rank : 151) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.014150 (rank : 143) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.030258 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.010905 (rank : 152) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.009092 (rank : 155) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
DYDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.014438 (rank : 142) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWB3, Q5QP03, Q5QP04, Q6WNP4, Q6ZU87 | Gene names | DYDC1, DPY30D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 1 (RSD-9). | |||||
|
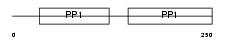
FETUB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.013833 (rank : 144) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |
|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
GCP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.021347 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.012228 (rank : 148) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.003976 (rank : 168) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.006106 (rank : 164) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.005782 (rank : 165) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.019841 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.008098 (rank : 159) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.008262 (rank : 158) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.006906 (rank : 161) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.006859 (rank : 162) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.014657 (rank : 140) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.011140 (rank : 150) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.010088 (rank : 153) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.082082 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.082145 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.053811 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.051449 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052999 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
E41L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.090186 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.089416 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.083224 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.079978 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.084749 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.082434 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.099160 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
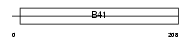
E41LA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.096977 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.096956 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
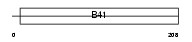
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.092860 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
E41LB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.095377 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.104263 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.104861 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.081484 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.080578 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.073975 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FRMD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.065877 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.104905 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.104137 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.112826 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
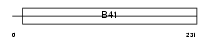
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.102329 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
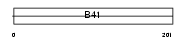
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
MERL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.111118 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.110302 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MOES_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.099515 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.097813 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052646 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.091627 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.096013 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.052401 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.058961 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.050131 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.057597 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PTN13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.056042 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.056960 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.068393 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.068801 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.068884 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.070092 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.059669 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.059176 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
RADI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.104331 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.103422 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.052393 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.965510 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.479261 (rank : 3) | θ value | 4.72714e-33 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.478481 (rank : 4) | θ value | 4.00176e-32 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.438201 (rank : 5) | θ value | 2.69047e-20 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.431890 (rank : 6) | θ value | 2.27762e-19 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
THSD1_MOUSE
|
||||||
| NC score | 0.116539 (rank : 7) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
FRMD6_HUMAN
|
||||||
| NC score | 0.112826 (rank : 8) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.111118 (rank : 9) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.110302 (rank : 10) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.104905 (rank : 11) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.104861 (rank : 12) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.104331 (rank : 13) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.104263 (rank : 14) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FRMD5_MOUSE
|
||||||
| NC score | 0.104137 (rank : 15) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.103422 (rank : 16) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.102329 (rank : 17) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.099515 (rank : 18) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
E41L5_HUMAN
|
||||||
| NC score | 0.099160 (rank : 19) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.097813 (rank : 20) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
E41LA_HUMAN
|
||||||
| NC score | 0.096977 (rank : 21) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| NC score | 0.096956 (rank : 22) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.096013 (rank : 23) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
E41LB_MOUSE
|
||||||
| NC score | 0.095377 (rank : 24) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.092860 (rank : 25) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.091627 (rank : 26) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.091220 (rank : 27) | θ value | 7.1131e-05 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.090186 (rank : 28) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.089416 (rank : 29) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.084749 (rank : 30) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.083224 (rank : 31) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.082434 (rank : 32) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.082145 (rank : 33) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.082082 (rank : 34) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.081484 (rank : 35) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.080578 (rank : 36) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.079978 (rank : 37) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.077638 (rank : 38) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.073975 (rank : 39) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.071939 (rank : 40) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.071484 (rank : 41) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.070092 (rank : 42) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.069920 (rank : 43) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.068884 (rank : 44) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.068801 (rank : 45) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.068741 (rank : 46) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.068466 (rank : 47) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.068393 (rank : 48) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.067770 (rank : 49) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.067655 (rank : 50) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
FRMD1_HUMAN
|
||||||
| NC score | 0.065877 (rank : 51) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.063133 (rank : 52) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.062780 (rank : 53) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.061964 (rank : 54) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.061227 (rank : 55) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.061109 (rank : 56) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.059669 (rank : 57) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.059603 (rank : 58) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.059176 (rank : 59) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.058961 (rank : 60) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.057597 (rank : 61) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.056960 (rank : 62) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.056042 (rank : 63) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.054878 (rank : 64) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.053811 (rank : 65) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.052999 (rank : 66) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.052653 (rank : 67) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.052646 (rank : 68) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.052401 (rank : 69) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.052393 (rank : 70) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.051449 (rank : 71) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.050168 (rank : 72) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.050131 (rank : 73) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.048468 (rank : 74) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
MUC15_HUMAN
|
||||||
| NC score | 0.047516 (rank : 75) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.044701 (rank : 76) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.043158 (rank : 77) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.041834 (rank : 78) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CU056_HUMAN
|
||||||
| NC score | 0.040596 (rank : 79) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.039453 (rank : 80) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.039215 (rank : 81) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.039183 (rank : 82) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.037891 (rank : 83) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
ENK10_HUMAN
|
||||||
| NC score | 0.037434 (rank : 84) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10267, Q14273 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Env polyprotein (Envelope polyprotein) (HERV-K10 envelope protein) (HERV-K107 envelope protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK11_HUMAN
|
||||||
| NC score | 0.037431 (rank : 85) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61566 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Env polyprotein (Envelope polyprotein) (HERV-K101 envelope protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK12_HUMAN
|
||||||
| NC score | 0.037393 (rank : 86) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61567 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Env polyprotein (Envelope polyprotein) (HERV-K102 envelope protein) (HERV-K(III) envelope protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
PP4R1_MOUSE
|
||||||
| NC score | 0.036775 (rank : 87) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.035894 (rank : 88) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.035446 (rank : 89) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CCD24_HUMAN
|
||||||
| NC score | 0.035376 (rank : 90) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N4L8, Q6RWT2 | Gene names | CCDC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 24. | |||||
|
ENK1_HUMAN
|
||||||
| NC score | 0.035055 (rank : 91) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61565 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (EnvK1 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK4_HUMAN
|
||||||
| NC score | 0.034921 (rank : 92) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKH3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_6q14.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K109 envelope protein) (HERV-K(C6) envelope protein) (EnvK4 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK2_HUMAN
|
||||||
| NC score | 0.034888 (rank : 93) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q69384, Q8TD93, Q9UBU4, Q9YNA5, Q9YNA9 | Gene names | ERVK6 | |||
|
Domain Architecture |

|
|||||
| Description | HERV-K_7p22.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K(HML-2.HOM) envelope protein) (HERV-K108 envelope protein) (HERV-K(C7) envelope protein) (EnvK2 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK6_HUMAN
|
||||||
| NC score | 0.034859 (rank : 94) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q902F8 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_8p23.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K115 envelope protein) (EnvK6 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK3_HUMAN
|
||||||
| NC score | 0.034849 (rank : 95) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O71037, Q69386, Q9YNA7 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_19q12 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-K(C19) envelope protein) (EnvK3 protein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
ENK17_HUMAN
|
||||||
| NC score | 0.034742 (rank : 96) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P61570 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Env polyprotein precursor (Envelope polyprotein) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.034673 (rank : 97) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.034647 (rank : 98) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.033249 (rank : 99) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.033050 (rank : 100) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.031980 (rank : 101) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
PABP2_MOUSE
|
||||||
| NC score | 0.031530 (rank : 102) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CCS6, O35935 | Gene names | Pabpn1, Pab2, Pabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
PABP2_HUMAN
|
||||||
| NC score | 0.031168 (rank : 103) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86U42, O43484 | Gene names | PABPN1, PAB2, PABP2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.030970 (rank : 104) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.030961 (rank : 105) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.030870 (rank : 106) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.030564 (rank : 107) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.030258 (rank : 108) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
SAKS1_HUMAN
|
||||||
| NC score | 0.029529 (rank : 109) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
POK12_HUMAN
|
||||||
| NC score | 0.028360 (rank : 110) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63135 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pol protein (HERV-K102 Pol protein) (HERV-K(III) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.028116 (rank : 111) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.026886 (rank : 112) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.026099 (rank : 113) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
MLH1_HUMAN
|
||||||
| NC score | 0.025942 (rank : 114) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
WDR67_MOUSE
|
||||||
| NC score | 0.025723 (rank : 115) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NXY1, Q3TBW4, Q3U4F6, Q5KSA3, Q810J7, Q8C3Y2 | Gene names | Wdr67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67 (Protein 4-B-3). | |||||
|
N4BP3_HUMAN
|
||||||
| NC score | 0.025475 (rank : 116) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15049, Q7Z6I3 | Gene names | N4BP3, KIAA0341 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
DTNB_MOUSE
|
||||||
| NC score | 0.025399 (rank : 117) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.023506 (rank : 118) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.023354 (rank : 119) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.022668 (rank : 120) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
NAB2_MOUSE
|
||||||
| NC score | 0.022293 (rank : 121) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.022254 (rank : 122) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.021945 (rank : 123) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
GCP6_HUMAN
|
||||||
| NC score | 0.021347 (rank : 124) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.021081 (rank : 125) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.020959 (rank : 126) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.020727 (rank : 127) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.019841 (rank : 128) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.019427 (rank : 129) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.019263 (rank : 130) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
SP100_MOUSE
|
||||||
| NC score | 0.019005 (rank : 131) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35892, O35897, O88392, O88395 | Gene names | Sp100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.018749 (rank : 132) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.018138 (rank : 133) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.017933 (rank : 134) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.017914 (rank : 135) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.017666 (rank : 136) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.015810 (rank : 137) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
BCN1_MOUSE
|
||||||
| NC score | 0.015754 (rank : 138) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88597 | Gene names | Becn1 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein). | |||||
|
FOXGA_HUMAN
|
||||||
| NC score | 0.014892 (rank : 139) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55316 | Gene names | FOXG1A, FKHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1A (Forkhead-related protein FKHL2) (Transcription factor BF-2) (Brain factor 2) (BF2) (HFK2). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.014657 (rank : 140) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.014500 (rank : 141) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DYDC1_HUMAN
|
||||||
| NC score | 0.014438 (rank : 142) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWB3, Q5QP03, Q5QP04, Q6WNP4, Q6ZU87 | Gene names | DYDC1, DPY30D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 1 (RSD-9). | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.014150 (rank : 143) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
FETUB_HUMAN
|
||||||
| NC score | 0.013833 (rank : 144) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |

|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.013646 (rank : 145) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ATS12_HUMAN
|
||||||
| NC score | 0.012760 (rank : 146) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.012612 (rank : 147) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.012228 (rank : 148) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.011574 (rank : 149) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.011140 (rank : 150) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
RAVR2_MOUSE
|
||||||
| NC score | 0.011028 (rank : 151) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.010905 (rank : 152) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.010088 (rank : 153) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.009375 (rank : 154) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.009092 (rank : 155) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.008607 (rank : 156) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
PO2F2_HUMAN
|
||||||
| NC score | 0.008422 (rank : 157) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.008262 (rank : 158) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.008098 (rank : 159) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.007906 (rank : 160) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.006906 (rank : 161) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.006859 (rank : 162) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
PRGR_MOUSE
|
||||||
| NC score | 0.006114 (rank : 163) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.006106 (rank : 164) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.005782 (rank : 165) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
STK36_HUMAN
|
||||||
| NC score | 0.005708 (rank : 166) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
KSYK_MOUSE
|
||||||
| NC score | 0.004497 (rank : 167) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.003976 (rank : 168) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.003429 (rank : 169) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
HIC1_MOUSE
|
||||||
| NC score | 0.002500 (rank : 170) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.002426 (rank : 171) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
ZN250_MOUSE
|
||||||
| NC score | -0.001109 (rank : 172) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNU6, Q8BJ57 | Gene names | Znf250, Zfp647, Znf647 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 250 (Zinc finger protein 647). | |||||