Please be patient as the page loads
|
E41LA_HUMAN
|
||||||
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |
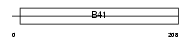
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
E41LA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996444 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |
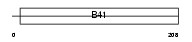
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41L5_HUMAN
|
||||||
| θ value | 5.33382e-77 (rank : 3) | NC score | 0.972152 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
E41LB_MOUSE
|
||||||
| θ value | 3.45727e-76 (rank : 4) | NC score | 0.972385 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 5) | NC score | 0.967006 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 2.47765e-74 (rank : 6) | NC score | 0.680963 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
E41L3_HUMAN
|
||||||
| θ value | 3.70236e-70 (rank : 7) | NC score | 0.958406 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L1_MOUSE
|
||||||
| θ value | 1.40689e-69 (rank : 8) | NC score | 0.956585 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 4.09345e-69 (rank : 9) | NC score | 0.952151 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 1.5555e-68 (rank : 10) | NC score | 0.945135 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
FRMD5_MOUSE
|
||||||
| θ value | 2.65329e-68 (rank : 11) | NC score | 0.970542 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
41_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 12) | NC score | 0.953568 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | 2.93355e-67 (rank : 13) | NC score | 0.968595 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
41_MOUSE
|
||||||
| θ value | 2.74572e-65 (rank : 14) | NC score | 0.950157 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 3.58603e-65 (rank : 15) | NC score | 0.940362 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 2.84138e-62 (rank : 16) | NC score | 0.918005 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | 2.1121e-49 (rank : 17) | NC score | 0.653753 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 18) | NC score | 0.788577 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 5.95217e-44 (rank : 19) | NC score | 0.756131 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 7.27602e-42 (rank : 20) | NC score | 0.763844 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 21) | NC score | 0.627659 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 22) | NC score | 0.625575 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 23) | NC score | 0.751994 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 24) | NC score | 0.747338 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 25) | NC score | 0.612848 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 26) | NC score | 0.733382 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 27) | NC score | 0.729733 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 28) | NC score | 0.689017 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 8.3442e-30 (rank : 29) | NC score | 0.681432 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 30) | NC score | 0.604937 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 1.02001e-27 (rank : 31) | NC score | 0.788070 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 1.33217e-27 (rank : 32) | NC score | 0.788786 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 33) | NC score | 0.489529 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 34) | NC score | 0.498954 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 35) | NC score | 0.846554 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 36) | NC score | 0.862704 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 37) | NC score | 0.805887 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 7.56453e-23 (rank : 38) | NC score | 0.804877 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 39) | NC score | 0.808257 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 40) | NC score | 0.810874 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRMD6_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 41) | NC score | 0.700185 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |
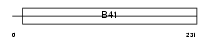
|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 42) | NC score | 0.706082 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |
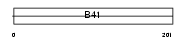
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
TLN2_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 43) | NC score | 0.441439 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
TLN2_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 44) | NC score | 0.446148 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 45) | NC score | 0.447588 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 46) | NC score | 0.444518 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
FRMD1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 47) | NC score | 0.674434 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 48) | NC score | 0.031576 (rank : 133) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 49) | NC score | 0.040706 (rank : 125) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
WAPL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 50) | NC score | 0.047249 (rank : 122) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.029512 (rank : 134) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 52) | NC score | 0.052134 (rank : 119) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.054037 (rank : 117) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 54) | NC score | 0.051307 (rank : 120) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
WAPL_MOUSE
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.047144 (rank : 123) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.014524 (rank : 144) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
FUSIP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.019180 (rank : 141) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |
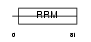
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.019180 (rank : 142) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |
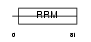
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.039506 (rank : 126) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.039462 (rank : 127) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.034702 (rank : 130) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.026235 (rank : 136) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.027257 (rank : 135) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.020783 (rank : 140) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.022024 (rank : 137) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.035629 (rank : 129) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.032448 (rank : 132) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.044639 (rank : 124) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.038257 (rank : 128) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WDHD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.014055 (rank : 145) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
SRPK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.001104 (rank : 156) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.033412 (rank : 131) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.010541 (rank : 148) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.015692 (rank : 143) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.009402 (rank : 150) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.021342 (rank : 138) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
KBTBA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.004262 (rank : 153) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60662 | Gene names | KBTBD10, KRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 10 (Kelch-related protein 1) (Kel-like protein 23) (Sarcosin). | |||||
|
STOX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.021013 (rank : 139) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.007087 (rank : 152) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ABCA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.001159 (rank : 155) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35600 | Gene names | Abca4, Abcr | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP). | |||||
|
AKT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | -0.004883 (rank : 158) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31751, Q68GC0 | Gene names | AKT2 | |||
|
Domain Architecture |
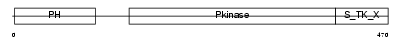
|
|||||
| Description | RAC-beta serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-beta) (Protein kinase Akt-2) (Protein kinase B, beta) (PKB beta). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.009891 (rank : 149) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.007722 (rank : 151) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.012988 (rank : 147) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.013971 (rank : 146) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SPG16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | -0.001108 (rank : 157) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N0X2, Q658W1, Q68DB3, Q6I9Z6, Q8N9C7, Q9H601 | Gene names | SPAG16, PF20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 16 protein (PF20 protein homolog). | |||||
|
ZN512_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.001190 (rank : 154) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96ME7, Q96JM0 | Gene names | ZNF512, KIAA1805 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 512. | |||||
|
CA106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.096977 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.095638 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
CD45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.105499 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CD45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.110104 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.054663 (rank : 116) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.052267 (rank : 118) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054986 (rank : 115) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.055456 (rank : 113) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.058411 (rank : 109) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.054994 (rank : 114) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
FGD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.056949 (rank : 111) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.057140 (rank : 110) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.059935 (rank : 107) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.055479 (rank : 112) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.059072 (rank : 108) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
KRIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.091640 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |
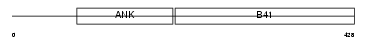
|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
KRIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.084862 (rank : 100) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
MESD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.061787 (rank : 106) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1K6 | Gene names | MESDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
MESD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.063528 (rank : 105) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERE8, Q542D0 | Gene names | Mesdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.051131 (rank : 121) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PTN11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.109511 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.109518 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTN12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.116522 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.116510 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTN18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.125965 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.122098 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.125843 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.126236 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTN22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.119530 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.117665 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.109529 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.108537 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.125848 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.127180 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.109458 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.109414 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.116421 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.116403 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTN7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.116660 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTN7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.117916 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTN9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.121559 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.121176 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTPR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.113606 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.113814 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.101649 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.101533 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.106955 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.071029 (rank : 102) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.068347 (rank : 104) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.102065 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.101537 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.069143 (rank : 103) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.090731 (rank : 94) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.090759 (rank : 93) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.109127 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.109565 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.091542 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.092324 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.086140 (rank : 99) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.086553 (rank : 97) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.113789 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.113550 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.114060 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.111536 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
PTPRR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.111874 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.072257 (rank : 101) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.088669 (rank : 95) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.088531 (rank : 96) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
PTPRU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.086210 (rank : 98) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.108495 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.091313 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
E41LA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| NC score | 0.996444 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LB_MOUSE
|
||||||
| NC score | 0.972385 (rank : 3) | θ value | 3.45727e-76 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
E41L5_HUMAN
|
||||||
| NC score | 0.972152 (rank : 4) | θ value | 5.33382e-77 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
FRMD5_MOUSE
|
||||||
| NC score | 0.970542 (rank : 5) | θ value | 2.65329e-68 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.968595 (rank : 6) | θ value | 2.93355e-67 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.967006 (rank : 7) | θ value | 2.92676e-75 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.958406 (rank : 8) | θ value | 3.70236e-70 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.956585 (rank : 9) | θ value | 1.40689e-69 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.953568 (rank : 10) | θ value | 7.71989e-68 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.952151 (rank : 11) | θ value | 4.09345e-69 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.950157 (rank : 12) | θ value | 2.74572e-65 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.945135 (rank : 13) | θ value | 1.5555e-68 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.940362 (rank : 14) | θ value | 3.58603e-65 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.918005 (rank : 15) | θ value | 2.84138e-62 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.862704 (rank : 16) | θ value | 1.16704e-23 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.846554 (rank : 17) | θ value | 1.16704e-23 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.810874 (rank : 18) | θ value | 1.2077e-20 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.808257 (rank : 19) | θ value | 9.24701e-21 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.805887 (rank : 20) | θ value | 4.43474e-23 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.804877 (rank : 21) | θ value | 7.56453e-23 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.788786 (rank : 22) | θ value | 1.33217e-27 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.788577 (rank : 23) | θ value | 1.51363e-47 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.788070 (rank : 24) | θ value | 1.02001e-27 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.763844 (rank : 25) | θ value | 7.27602e-42 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.756131 (rank : 26) | θ value | 5.95217e-44 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.751994 (rank : 27) | θ value | 1.98606e-31 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.747338 (rank : 28) | θ value | 1.98606e-31 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.733382 (rank : 29) | θ value | 9.8567e-31 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.729733 (rank : 30) | θ value | 9.8567e-31 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.706082 (rank : 31) | θ value | 4.59992e-12 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
FRMD6_HUMAN
|
||||||
| NC score | 0.700185 (rank : 32) | θ value | 7.09661e-13 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.689017 (rank : 33) | θ value | 8.3442e-30 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.681432 (rank : 34) | θ value | 8.3442e-30 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.680963 (rank : 35) | θ value | 2.47765e-74 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
FRMD1_HUMAN
|
||||||
| NC score | 0.674434 (rank : 36) | θ value | 1.53129e-07 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.653753 (rank : 37) | θ value | 2.1121e-49 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.627659 (rank : 38) | θ value | 2.77131e-33 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.625575 (rank : 39) | θ value | 1.79631e-32 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.612848 (rank : 40) | θ value | 5.77852e-31 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.604937 (rank : 41) | θ value | 1.85889e-29 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.498954 (rank : 42) | θ value | 8.63488e-27 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.489529 (rank : 43) | θ value | 2.27234e-27 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.447588 (rank : 44) | θ value | 1.63604e-09 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN2_MOUSE
|
||||||
| NC score | 0.446148 (rank : 45) | θ value | 1.25267e-09 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.444518 (rank : 46) | θ value | 1.63604e-09 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN2_HUMAN
|
||||||
| NC score | 0.441439 (rank : 47) | θ value | 9.59137e-10 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
PTN2_MOUSE
|
||||||
| NC score | 0.127180 (rank : 48) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN1_MOUSE
|
||||||
| NC score | 0.126236 (rank : 49) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTN18_HUMAN
|
||||||
| NC score | 0.125965 (rank : 50) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN2_HUMAN
|
||||||
| NC score | 0.125848 (rank : 51) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTN1_HUMAN
|
||||||
| NC score | 0.125843 (rank : 52) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.122098 (rank : 53) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN9_HUMAN
|
||||||
| NC score | 0.121559 (rank : 54) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| NC score | 0.121176 (rank : 55) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.119530 (rank : 56) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN7_MOUSE
|
||||||
| NC score | 0.117916 (rank : 57) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.117665 (rank : 58) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN7_HUMAN
|
||||||
| NC score | 0.116660 (rank : 59) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.116522 (rank : 60) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.116510 (rank : 61) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTN6_HUMAN
|
||||||
| NC score | 0.116421 (rank : 62) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| NC score | 0.116403 (rank : 63) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.114060 (rank : 64) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPR2_MOUSE
|
||||||
| NC score | 0.113814 (rank : 65) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.113789 (rank : 66) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPR2_HUMAN
|
||||||
| NC score | 0.113606 (rank : 67) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.113550 (rank : 68) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRR_MOUSE
|
||||||
| NC score | 0.111874 (rank : 69) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.111536 (rank : 70) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.110104 (rank : 71) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.109565 (rank : 72) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.109529 (rank : 73) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN11_MOUSE
|
||||||
| NC score | 0.109518 (rank : 74) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_HUMAN
|
||||||
| NC score | 0.109511 (rank : 75) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN5_HUMAN
|
||||||
| NC score | 0.109458 (rank : 76) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.109414 (rank : 77) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.109127 (rank : 78) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.108537 (rank : 79) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.108495 (rank : 80) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTPRB_HUMAN
|
||||||
| NC score | 0.106955 (rank : 81) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.105499 (rank : 82) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
PTPRE_HUMAN
|
||||||
| NC score | 0.102065 (rank : 83) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.101649 (rank : 84) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRE_MOUSE
|
||||||
| NC score | 0.101537 (rank : 85) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRA_MOUSE
|
||||||
| NC score | 0.101533 (rank : 86) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.096977 (rank : 87) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.095638 (rank : 88) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.092324 (rank : 89) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
KRIT1_HUMAN
|
||||||
| NC score | 0.091640 (rank : 90) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |

|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.091542 (rank : 91) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.091313 (rank : 92) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.090759 (rank : 93) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.090731 (rank : 94) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRT_HUMAN
|
||||||
| NC score | 0.088669 (rank : 95) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.088531 (rank : 96) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
PTPRM_MOUSE
|
||||||
| NC score | 0.086553 (rank : 97) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRU_HUMAN
|
||||||
| NC score | 0.086210 (rank : 98) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
PTPRM_HUMAN
|
||||||
| NC score | 0.086140 (rank : 99) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
KRIT1_MOUSE
|
||||||
| NC score | 0.084862 (rank : 100) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
PTPRS_HUMAN
|
||||||
| NC score | 0.072257 (rank : 101) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.071029 (rank : 102) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRF_HUMAN
|
||||||
| NC score | 0.069143 (rank : 103) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.068347 (rank : 104) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
MESD1_MOUSE
|
||||||
| NC score | 0.063528 (rank : 105) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERE8, Q542D0 | Gene names | Mesdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
MESD1_HUMAN
|
||||||
| NC score | 0.061787 (rank : 106) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1K6 | Gene names | MESDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.059935 (rank : 107) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.059072 (rank : 108) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.058411 (rank : 109) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.057140 (rank : 110) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.056949 (rank : 111) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.055479 (rank : 112) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.055456 (rank : 113) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.054994 (rank : 114) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.054986 (rank : 115) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.054663 (rank : 116) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.054037 (rank : 117) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.052267 (rank : 118) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.052134 (rank : 119) | θ value | 0.0736092 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.051307 (rank : 120) | θ value | 0.125558 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.051131 (rank : 121) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
WAPL_HUMAN
|
||||||
| NC score | 0.047249 (rank : 122) | θ value | 0.0431538 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
WAPL_MOUSE
|
||||||
| NC score | 0.047144 (rank : 123) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.044639 (rank : 124) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.040706 (rank : 125) | θ value | 0.0330416 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.039506 (rank : 126) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.039462 (rank : 127) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.038257 (rank : 128) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.035629 (rank : 129) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.034702 (rank : 130) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.033412 (rank : 131) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.032448 (rank : 132) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.031576 (rank : 133) | θ value | 0.0113563 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.029512 (rank : 134) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.027257 (rank : 135) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.026235 (rank : 136) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.022024 (rank : 137) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.021342 (rank : 138) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
STOX1_HUMAN
|
||||||
| NC score | 0.021013 (rank : 139) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.020783 (rank : 140) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.019180 (rank : 141) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.019180 (rank : 142) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.015692 (rank : 143) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.014524 (rank : 144) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
WDHD1_MOUSE
|
||||||
| NC score | 0.014055 (rank : 145) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.013971 (rank : 146) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.012988 (rank : 147) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.010541 (rank : 148) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.009891 (rank : 149) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.009402 (rank : 150) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.007722 (rank : 151) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.007087 (rank : 152) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
KBTBA_HUMAN
|
||||||
| NC score | 0.004262 (rank : 153) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60662 | Gene names | KBTBD10, KRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 10 (Kelch-related protein 1) (Kel-like protein 23) (Sarcosin). | |||||
|
ZN512_HUMAN
|
||||||
| NC score | 0.001190 (rank : 154) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96ME7, Q96JM0 | Gene names | ZNF512, KIAA1805 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 512. | |||||
|
ABCA4_MOUSE
|
||||||
| NC score | 0.001159 (rank : 155) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35600 | Gene names | Abca4, Abcr | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP). | |||||
|
SRPK1_MOUSE
|
||||||
| NC score | 0.001104 (rank : 156) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
SPG16_HUMAN
|
||||||
| NC score | -0.001108 (rank : 157) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N0X2, Q658W1, Q68DB3, Q6I9Z6, Q8N9C7, Q9H601 | Gene names | SPAG16, PF20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 16 protein (PF20 protein homolog). | |||||
|
AKT2_HUMAN
|
||||||
| NC score | -0.004883 (rank : 158) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31751, Q68GC0 | Gene names | AKT2 | |||
|
Domain Architecture |

|
|||||
| Description | RAC-beta serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-beta) (Protein kinase Akt-2) (Protein kinase B, beta) (PKB beta). | |||||