Please be patient as the page loads
|
FRM4B_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FRM4A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.981749 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982395 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996374 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 5) | NC score | 0.725091 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | 6.59618e-35 (rank : 6) | NC score | 0.721880 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 7) | NC score | 0.736582 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 8) | NC score | 0.740215 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 9) | NC score | 0.683581 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 8.06329e-33 (rank : 10) | NC score | 0.675276 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 11) | NC score | 0.770844 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 12) | NC score | 0.770998 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 13) | NC score | 0.790524 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 14) | NC score | 0.794102 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 15) | NC score | 0.819758 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 16) | NC score | 0.819853 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 17) | NC score | 0.775304 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L5_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 18) | NC score | 0.811056 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 19) | NC score | 0.564452 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 20) | NC score | 0.553558 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
E41L3_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 21) | NC score | 0.789774 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 22) | NC score | 0.561609 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 23) | NC score | 0.546899 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
E41LB_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 24) | NC score | 0.805023 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 25) | NC score | 0.776700 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 26) | NC score | 0.751844 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41LA_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 27) | NC score | 0.810874 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 28) | NC score | 0.649467 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 29) | NC score | 0.796401 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 30) | NC score | 0.555067 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 31) | NC score | 0.564782 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
41_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 32) | NC score | 0.782096 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
CA106_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 33) | NC score | 0.431890 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 34) | NC score | 0.666969 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
E41LA_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 35) | NC score | 0.809798 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
41_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 36) | NC score | 0.776242 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
CA106_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 37) | NC score | 0.428329 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 38) | NC score | 0.638033 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 39) | NC score | 0.444170 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 40) | NC score | 0.452752 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 41) | NC score | 0.763904 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 42) | NC score | 0.746546 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
FRMD6_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 43) | NC score | 0.699692 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 44) | NC score | 0.694254 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
FRMD1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 45) | NC score | 0.640064 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
TLN2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 46) | NC score | 0.367989 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
TLN2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 47) | NC score | 0.361591 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 48) | NC score | 0.062423 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 49) | NC score | 0.363024 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 50) | NC score | 0.360735 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.045354 (rank : 149) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
KRIT1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 52) | NC score | 0.123639 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |
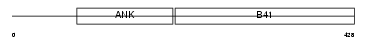
|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.054660 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 54) | NC score | 0.042315 (rank : 157) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.026667 (rank : 173) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.045748 (rank : 148) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.039723 (rank : 159) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.044150 (rank : 151) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.016908 (rank : 180) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.043043 (rank : 155) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.055094 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.043144 (rank : 154) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
CCD38_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.035783 (rank : 164) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.054622 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
MCES_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.036580 (rank : 163) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
PESC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.040389 (rank : 158) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
THAP6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.032937 (rank : 167) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |
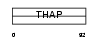
|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.000425 (rank : 193) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZO2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.028358 (rank : 171) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.049450 (rank : 140) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.047841 (rank : 143) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.047487 (rank : 144) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.037877 (rank : 160) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
KRIT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.104694 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
NCOA7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.014264 (rank : 182) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
WDR67_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.056314 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
CCD13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.049609 (rank : 139) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CN145_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.048846 (rank : 141) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.049802 (rank : 138) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.005624 (rank : 189) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
TRI59_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.014282 (rank : 181) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q922Y2, Q9CSP2, Q9CUD5, Q9D740 | Gene names | Trim59, Mrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (RING finger protein 1). | |||||
|
ATS5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.005136 (rank : 191) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA0, Q9UKP2 | Gene names | ADAMTS5, ADAMTS11, ADMP2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-5 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 5) (ADAM-TS 5) (ADAM-TS5) (Aggrecanase-2) (ADMP-2) (ADAM-TS 11). | |||||
|
CF182_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.043668 (rank : 152) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
KINH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.031182 (rank : 168) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.032974 (rank : 166) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NADAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.017388 (rank : 179) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.008470 (rank : 186) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PAK7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | -0.002070 (rank : 194) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||
|
SPG21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.026535 (rank : 174) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZD8, Q6ZMB6 | Gene names | SPG21, ACP33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maspardin (Spastic paraplegia 21 autosomal recessive Mast syndrome protein) (Acid cluster protein 33). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.028708 (rank : 170) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.021789 (rank : 178) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.010891 (rank : 184) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.043241 (rank : 153) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.029472 (rank : 169) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.047071 (rank : 146) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
PAK7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | -0.003321 (rank : 195) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P286, Q5W115, Q9BX09, Q9ULF6 | Gene names | PAK7, KIAA1264, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (PAK-5). | |||||
|
SPG21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.024429 (rank : 175) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQC8, Q3U658, Q3UDT7, Q8C1Y7 | Gene names | Spg21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maspardin (Spastic paraplegia 21 autosomal recessive Mast syndrome protein homolog) (Acid cluster protein 33). | |||||
|
ATPG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.009940 (rank : 185) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36542, Q5VYP3, Q96AS8 | Gene names | ATP5C1, ATP5C | |||
|
Domain Architecture |
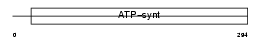
|
|||||
| Description | ATP synthase gamma chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
ATS5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.003893 (rank : 192) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R001 | Gene names | Adamts5 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-5 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 5) (ADAM-TS 5) (ADAM-TS5) (Aggrecanase-2) (ADMP-2) (Implantin). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.027671 (rank : 172) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.048197 (rank : 142) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
PKHC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.037449 (rank : 161) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.005143 (rank : 190) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.007357 (rank : 187) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
CENPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.050884 (rank : 136) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H3R5 | Gene names | CENPH, ICEN35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein H (CENP-H) (Interphase centromere complex protein 35). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.012839 (rank : 183) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CN145_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.042824 (rank : 156) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.044251 (rank : 150) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.047047 (rank : 147) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.022470 (rank : 177) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.023211 (rank : 176) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.005913 (rank : 188) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
PKHC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.033711 (rank : 165) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.047167 (rank : 145) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.058197 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.037076 (rank : 162) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZN583_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | -0.005820 (rank : 196) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V080, Q3V3L4 | Gene names | Znf583, Zfp583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583. | |||||
|
CCD11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.063063 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.060887 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IVM0, Q86VH7 | Gene names | CCDC50, C3orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.055520 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CD45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.087454 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CD45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.090585 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
CDC37_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.054813 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CDC37_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.050948 (rank : 135) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |
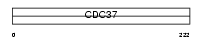
|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.053095 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CLCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053681 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.061770 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.062100 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.050200 (rank : 137) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.065976 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.061539 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.066713 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.064412 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054398 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.051979 (rank : 133) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PTN11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.090670 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.090661 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTN12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.096169 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.095960 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTN18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.103748 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.100556 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.103952 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.104263 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTN22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.097662 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.096381 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.090974 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.089572 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.103718 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.104924 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.090812 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.090766 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.096558 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.096463 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTN7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.096454 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTN7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.097438 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTN9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.100550 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.100228 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTPR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.092888 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.093020 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.084033 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.083861 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.088177 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.059014 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.056573 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.084180 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.083766 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.056654 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.074508 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.074520 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.089751 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.090126 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.075229 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.075895 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.070744 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.071117 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.093323 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.093288 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.093859 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.092306 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
PTPRR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.092653 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.059291 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.072876 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.072745 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
PTPRU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.070719 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.089500 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.075169 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.054082 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.056396 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.056445 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
SPC24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.058236 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
STXB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.055724 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.051745 (rank : 134) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.053218 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.059978 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.052197 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
ZWINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.055497 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.996374 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.982395 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.981749 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRMD5_MOUSE
|
||||||
| NC score | 0.819853 (rank : 5) | θ value | 4.43474e-23 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.819758 (rank : 6) | θ value | 1.5242e-23 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
E41L5_HUMAN
|
||||||
| NC score | 0.811056 (rank : 7) | θ value | 1.29031e-22 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
E41LA_HUMAN
|
||||||
| NC score | 0.810874 (rank : 8) | θ value | 1.2077e-20 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| NC score | 0.809798 (rank : 9) | θ value | 5.07402e-19 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LB_MOUSE
|
||||||
| NC score | 0.805023 (rank : 10) | θ value | 1.86321e-21 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.796401 (rank : 11) | θ value | 2.06002e-20 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.794102 (rank : 12) | θ value | 5.23862e-24 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.790524 (rank : 13) | θ value | 2.35151e-24 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.789774 (rank : 14) | θ value | 3.75424e-22 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.782096 (rank : 15) | θ value | 1.02238e-19 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.776700 (rank : 16) | θ value | 3.17815e-21 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.776242 (rank : 17) | θ value | 6.62687e-19 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.775304 (rank : 18) | θ value | 1.29031e-22 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.770998 (rank : 19) | θ value | 7.54701e-31 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.770844 (rank : 20) | θ value | 1.98606e-31 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.763904 (rank : 21) | θ value | 6.85773e-16 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.751844 (rank : 22) | θ value | 1.2077e-20 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.746546 (rank : 23) | θ value | 1.29331e-14 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.740215 (rank : 24) | θ value | 3.27365e-34 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.736582 (rank : 25) | θ value | 1.12514e-34 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.725091 (rank : 26) | θ value | 6.59618e-35 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.721880 (rank : 27) | θ value | 6.59618e-35 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
FRMD6_HUMAN
|
||||||
| NC score | 0.699692 (rank : 28) | θ value | 2.88119e-14 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.694254 (rank : 29) | θ value | 1.58096e-12 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.683581 (rank : 30) | θ value | 1.6247e-33 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.675276 (rank : 31) | θ value | 8.06329e-33 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.666969 (rank : 32) | θ value | 2.97466e-19 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.649467 (rank : 33) | θ value | 1.5773e-20 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FRMD1_HUMAN
|
||||||
| NC score | 0.640064 (rank : 34) | θ value | 4.92598e-06 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.638033 (rank : 35) | θ value | 1.24977e-17 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.564782 (rank : 36) | θ value | 3.51386e-20 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.564452 (rank : 37) | θ value | 2.20094e-22 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.561609 (rank : 38) | θ value | 3.75424e-22 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.555067 (rank : 39) | θ value | 2.69047e-20 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.553558 (rank : 40) | θ value | 2.87452e-22 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.546899 (rank : 41) | θ value | 8.36355e-22 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.452752 (rank : 42) | θ value | 1.058e-16 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.444170 (rank : 43) | θ value | 2.13179e-17 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.431890 (rank : 44) | θ value | 2.27762e-19 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.428329 (rank : 45) | θ value | 2.5182e-18 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
TLN2_MOUSE
|
||||||
| NC score | 0.367989 (rank : 46) | θ value | 0.000270298 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.363024 (rank : 47) | θ value | 0.00298849 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN2_HUMAN
|
||||||
| NC score | 0.361591 (rank : 48) | θ value | 0.000461057 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.360735 (rank : 49) | θ value | 0.00298849 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
KRIT1_HUMAN
|
||||||
| NC score | 0.123639 (rank : 50) | θ value | 0.125558 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |

|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
PTN2_MOUSE
|
||||||
| NC score | 0.104924 (rank : 51) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
KRIT1_MOUSE
|
||||||
| NC score | 0.104694 (rank : 52) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
PTN1_MOUSE
|
||||||
| NC score | 0.104263 (rank : 53) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTN1_HUMAN
|
||||||
| NC score | 0.103952 (rank : 54) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN18_HUMAN
|
||||||
| NC score | 0.103748 (rank : 55) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN2_HUMAN
|
||||||
| NC score | 0.103718 (rank : 56) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.100556 (rank : 57) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN9_HUMAN
|
||||||
| NC score | 0.100550 (rank : 58) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| NC score | 0.100228 (rank : 59) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.097662 (rank : 60) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN7_MOUSE
|
||||||
| NC score | 0.097438 (rank : 61) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTN6_HUMAN
|
||||||
| NC score | 0.096558 (rank : 62) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| NC score | 0.096463 (rank : 63) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTN7_HUMAN
|
||||||
| NC score | 0.096454 (rank : 64) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.096381 (rank : 65) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.096169 (rank : 66) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.095960 (rank : 67) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.093859 (rank : 68) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.093323 (rank : 69) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.093288 (rank : 70) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPR2_MOUSE
|
||||||
| NC score | 0.093020 (rank : 71) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPR2_HUMAN
|
||||||
| NC score | 0.092888 (rank : 72) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPRR_MOUSE
|
||||||
| NC score | 0.092653 (rank : 73) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.092306 (rank : 74) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.090974 (rank : 75) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN5_HUMAN
|
||||||
| NC score | 0.090812 (rank : 76) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.090766 (rank : 77) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN11_HUMAN
|
||||||
| NC score | 0.090670 (rank : 78) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_MOUSE
|
||||||
| NC score | 0.090661 (rank : 79) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.090585 (rank : 80) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.090126 (rank : 81) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.089751 (rank : 82) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.089572 (rank : 83) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.089500 (rank : 84) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTPRB_HUMAN
|
||||||
| NC score | 0.088177 (rank : 85) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.087454 (rank : 86) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
PTPRE_HUMAN
|
||||||
| NC score | 0.084180 (rank : 87) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.084033 (rank : 88) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| NC score | 0.083861 (rank : 89) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTPRE_MOUSE
|
||||||
| NC score | 0.083766 (rank : 90) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.075895 (rank : 91) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.075229 (rank : 92) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.075169 (rank : 93) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.074520 (rank : 94) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.074508 (rank : 95) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRT_HUMAN
|
||||||
| NC score | 0.072876 (rank : 96) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.072745 (rank : 97) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
PTPRM_MOUSE
|
||||||
| NC score | 0.071117 (rank : 98) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_HUMAN
|
||||||
| NC score | 0.070744 (rank : 99) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRU_HUMAN
|
||||||
| NC score | 0.070719 (rank : 100) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.066713 (rank : 101) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.065976 (rank : 102) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.064412 (rank : 103) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.063063 (rank : 104) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.062423 (rank : 105) | θ value | 0.000602161 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.062100 (rank : 106) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.061770 (rank : 107) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.061539 (rank : 108) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
CCD50_HUMAN
|
||||||
| NC score | 0.060887 (rank : 109) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IVM0, Q86VH7 | Gene names | CCDC50, C3orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.059978 (rank : 110) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
PTPRS_HUMAN
|
||||||
| NC score | 0.059291 (rank : 111) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.059014 (rank : 112) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
SPC24_HUMAN
|
||||||
| NC score | 0.058236 (rank : 113) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.058197 (rank : 114) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
PTPRF_HUMAN
|
||||||
| NC score | 0.056654 (rank : 115) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.056573 (rank : 116) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.056445 (rank : 117) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.056396 (rank : 118) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
WDR67_HUMAN
|
||||||
| NC score | 0.056314 (rank : 119) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.055724 (rank : 120) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.055520 (rank : 121) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
ZWINT_MOUSE
|
||||||
| NC score | 0.055497 (rank : 122) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.055094 (rank : 123) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.054813 (rank : 124) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.054660 (rank : 125) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.054622 (rank : 126) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.054398 (rank : 127) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.054082 (rank : 128) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
CLCB_MOUSE
|
||||||
| NC score | 0.053681 (rank : 129) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.053218 (rank : 130) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.053095 (rank : 131) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.052197 (rank : 132) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.051979 (rank : 133) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.051745 (rank : 134) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CDC37_MOUSE
|
||||||
| NC score | 0.050948 (rank : 135) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CENPH_HUMAN
|
||||||
| NC score | 0.050884 (rank : 136) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H3R5 | Gene names | CENPH, ICEN35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein H (CENP-H) (Interphase centromere complex protein 35). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.050200 (rank : 137) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.049802 (rank : 138) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.049609 (rank : 139) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.049450 (rank : 140) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.048846 (rank : 141) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.048197 (rank : 142) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.047841 (rank : 143) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.047487 (rank : 144) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.047167 (rank : 145) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.047071 (rank : 146) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.047047 (rank : 147) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.045748 (rank : 148) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.045354 (rank : 149) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.044251 (rank : 150) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.044150 (rank : 151) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
CF182_MOUSE
|
||||||
| NC score | 0.043668 (rank : 152) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.043241 (rank : 153) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.043144 (rank : 154) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.043043 (rank : 155) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.042824 (rank : 156) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.042315 (rank : 157) | θ value | 0.21417 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PESC_MOUSE
|
||||||
| NC score | 0.040389 (rank : 158) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.039723 (rank : 159) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.037877 (rank : 160) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
PKHC1_HUMAN
|
||||||
| NC score | 0.037449 (rank : 161) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.037076 (rank : 162) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MCES_HUMAN
|
||||||
| NC score | 0.036580 (rank : 163) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
CCD38_HUMAN
|
||||||
| NC score | 0.035783 (rank : 164) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
PKHC1_MOUSE
|
||||||
| NC score | 0.033711 (rank : 165) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.032974 (rank : 166) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
THAP6_HUMAN
|
||||||
| NC score | 0.032937 (rank : 167) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.031182 (rank : 168) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.029472 (rank : 169) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.028708 (rank : 170) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.028358 (rank : 171) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.027671 (rank : 172) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.026667 (rank : 173) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
SPG21_HUMAN
|
||||||
| NC score | 0.026535 (rank : 174) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZD8, Q6ZMB6 | Gene names | SPG21, ACP33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maspardin (Spastic paraplegia 21 autosomal recessive Mast syndrome protein) (Acid cluster protein 33). | |||||
|
SPG21_MOUSE
|
||||||
| NC score | 0.024429 (rank : 175) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQC8, Q3U658, Q3UDT7, Q8C1Y7 | Gene names | Spg21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maspardin (Spastic paraplegia 21 autosomal recessive Mast syndrome protein homolog) (Acid cluster protein 33). | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.023211 (rank : 176) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.022470 (rank : 177) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.021789 (rank : 178) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
NADAP_HUMAN
|
||||||
| NC score | 0.017388 (rank : 179) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.016908 (rank : 180) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRI59_MOUSE
|
||||||
| NC score | 0.014282 (rank : 181) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q922Y2, Q9CSP2, Q9CUD5, Q9D740 | Gene names | Trim59, Mrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (RING finger protein 1). | |||||
|
NCOA7_MOUSE
|
||||||
| NC score | 0.014264 (rank : 182) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.012839 (rank : 183) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.010891 (rank : 184) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
ATPG_HUMAN
|
||||||
| NC score | 0.009940 (rank : 185) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36542, Q5VYP3, Q96AS8 | Gene names | ATP5C1, ATP5C | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase gamma chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.008470 (rank : 186) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.007357 (rank : 187) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.005913 (rank : 188) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.005624 (rank : 189) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.005143 (rank : 190) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
ATS5_HUMAN
|
||||||
| NC score | 0.005136 (rank : 191) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA0, Q9UKP2 | Gene names | ADAMTS5, ADAMTS11, ADMP2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-5 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 5) (ADAM-TS 5) (ADAM-TS5) (Aggrecanase-2) (ADMP-2) (ADAM-TS 11). | |||||
|
ATS5_MOUSE
|
||||||
| NC score | 0.003893 (rank : 192) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R001 | Gene names | Adamts5 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-5 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 5) (ADAM-TS 5) (ADAM-TS5) (Aggrecanase-2) (ADMP-2) (Implantin). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.000425 (rank : 193) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
PAK7_MOUSE
|
||||||
| NC score | -0.002070 (rank : 194) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||
|
PAK7_HUMAN
|
||||||
| NC score | -0.003321 (rank : 195) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P286, Q5W115, Q9BX09, Q9ULF6 | Gene names | PAK7, KIAA1264, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (PAK-5). | |||||
|
ZN583_MOUSE
|
||||||
| NC score | -0.005820 (rank : 196) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V080, Q3V3L4 | Gene names | Znf583, Zfp583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583. | |||||