Please be patient as the page loads
|
TLN2_HUMAN
|
||||||
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TLN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979773 (rank : 3) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978750 (rank : 4) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 157 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
TLN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996891 (rank : 2) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 5) | NC score | 0.276201 (rank : 48) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 6) | NC score | 0.331505 (rank : 39) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 7) | NC score | 0.295600 (rank : 45) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
MESD1_MOUSE
|
||||||
| θ value | 4.9032e-22 (rank : 8) | NC score | 0.574736 (rank : 5) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERE8, Q542D0 | Gene names | Mesdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
MESD1_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 9) | NC score | 0.563966 (rank : 6) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1K6 | Gene names | MESDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
RADI_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 10) | NC score | 0.407490 (rank : 19) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 11) | NC score | 0.406770 (rank : 20) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
MYO7A_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 12) | NC score | 0.132745 (rank : 61) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13402, P78427, Q13321, Q14785, Q92821, Q92822 | Gene names | MYO7A, USH1B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-7A (Myosin VIIa). | |||||
|
E41LA_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 13) | NC score | 0.448232 (rank : 7) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |
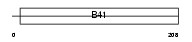
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
MYO7A_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 14) | NC score | 0.133263 (rank : 60) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97479 | Gene names | Myo7a, Myo7 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-7A (Myosin VIIa). | |||||
|
E41L5_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 15) | NC score | 0.437186 (rank : 9) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
PKHC1_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 16) | NC score | 0.345524 (rank : 35) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
PKHC1_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 17) | NC score | 0.343303 (rank : 36) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 18) | NC score | 0.434680 (rank : 12) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 19) | NC score | 0.415217 (rank : 15) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 20) | NC score | 0.417844 (rank : 14) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41LA_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 21) | NC score | 0.441439 (rank : 8) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |
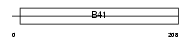
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 22) | NC score | 0.384029 (rank : 27) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 23) | NC score | 0.381100 (rank : 28) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 24) | NC score | 0.307406 (rank : 44) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
URP1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 25) | NC score | 0.333293 (rank : 38) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQL6, Q8IX34, Q8IYH2, Q9NWM2, Q9NXQ3 | Gene names | URP1, C20orf42, KIND1 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 1 (Kindlin-1) (Kindlerin) (Kindlin syndrome protein). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 26) | NC score | 0.406328 (rank : 21) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 27) | NC score | 0.405411 (rank : 22) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
URP2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 28) | NC score | 0.327734 (rank : 40) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UX7, Q8IUA1, Q8N207, Q9BT48 | Gene names | URP2, KIND3, MIG2B | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3) (MIG2-like). | |||||
|
URP2_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 29) | NC score | 0.326038 (rank : 41) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 30) | NC score | 0.434776 (rank : 11) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 31) | NC score | 0.435811 (rank : 10) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
E41LB_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 32) | NC score | 0.433099 (rank : 13) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 33) | NC score | 0.294508 (rank : 46) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
41_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 34) | NC score | 0.412886 (rank : 16) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 35) | NC score | 0.250974 (rank : 52) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
41_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 36) | NC score | 0.409333 (rank : 17) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
E41L3_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 37) | NC score | 0.408703 (rank : 18) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 38) | NC score | 0.399609 (rank : 25) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 39) | NC score | 0.237869 (rank : 56) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 40) | NC score | 0.397518 (rank : 26) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
KRIT1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 41) | NC score | 0.202410 (rank : 58) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |
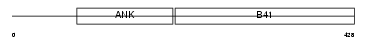
|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
KRIT1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 42) | NC score | 0.192427 (rank : 59) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
FRMD5_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 43) | NC score | 0.404292 (rank : 23) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 44) | NC score | 0.377497 (rank : 29) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 45) | NC score | 0.404119 (rank : 24) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 46) | NC score | 0.326036 (rank : 42) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |
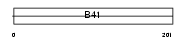
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 47) | NC score | 0.365201 (rank : 30) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 48) | NC score | 0.364777 (rank : 31) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 49) | NC score | 0.361591 (rank : 33) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRMD6_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 50) | NC score | 0.318498 (rank : 43) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 51) | NC score | 0.354471 (rank : 34) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 52) | NC score | 0.250047 (rank : 53) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 53) | NC score | 0.363703 (rank : 32) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
CLC4F_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 54) | NC score | 0.051723 (rank : 118) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 55) | NC score | 0.245496 (rank : 54) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
VINC_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 56) | NC score | 0.120404 (rank : 62) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18206, Q16450, Q5SWX2, Q7Z3B8, Q8IXU7 | Gene names | VCL | |||
|
Domain Architecture |
|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
VINC_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 57) | NC score | 0.117034 (rank : 63) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64727, Q8BP32, Q8BS46, Q922C5, Q922D9 | Gene names | Vcl | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 58) | NC score | 0.341566 (rank : 37) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 59) | NC score | 0.242628 (rank : 55) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.048752 (rank : 133) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
DOK4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.095194 (rank : 64) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 62) | NC score | 0.095041 (rank : 65) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 63) | NC score | 0.057780 (rank : 78) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.125558 (rank : 64) | NC score | 0.055333 (rank : 84) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 0.125558 (rank : 65) | NC score | 0.053468 (rank : 100) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 0.125558 (rank : 66) | NC score | 0.053404 (rank : 101) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 67) | NC score | 0.057392 (rank : 80) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 68) | NC score | 0.070101 (rank : 67) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | 0.041406 (rank : 144) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.027498 (rank : 159) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
M6PBP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 71) | NC score | 0.075910 (rank : 66) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60664, Q53G77, Q9BS03, Q9UBD7, Q9UP92 | Gene names | M6PRBP1, TIP47 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-6-phosphate receptor-binding protein 1 (Cargo selection protein TIP47) (47 kDa mannose 6-phosphate receptor-binding protein) (47 kDa MPR-binding protein) (Placental protein 17) (PP17). | |||||
|
DOK6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.053009 (rank : 110) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PKX4, Q4V9S3, Q8WUZ8, Q96HI2, Q96LU2 | Gene names | DOK6, DOK5L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 6 (Downstream of tyrosine kinase 6). | |||||
|
K1C16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.018782 (rank : 169) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08779, P30654, Q16402, Q9UBG8 | Gene names | KRT16, KRT16A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | 0.47712 (rank : 74) | NC score | 0.237136 (rank : 57) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 0.813845 (rank : 75) | NC score | 0.050456 (rank : 127) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.813845 (rank : 76) | NC score | 0.053309 (rank : 102) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.049986 (rank : 131) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.271969 (rank : 49) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.043517 (rank : 141) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.046311 (rank : 136) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.269036 (rank : 51) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.039469 (rank : 148) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.046744 (rank : 135) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
RNF6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.022442 (rank : 163) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.049393 (rank : 132) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CTNA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.058857 (rank : 77) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35221, Q12795 | Gene names | CTNNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (Cadherin-associated protein) (Alpha E-catenin) (NY- REN-13 antigen). | |||||
|
CTNA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.058891 (rank : 76) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26231 | Gene names | Ctnna1, Catna1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (102 kDa cadherin-associated protein) (CAP102) (Alpha E-catenin). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.280127 (rank : 47) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
ITA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 89) | NC score | 0.010338 (rank : 179) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
K0372_HUMAN
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.031064 (rank : 156) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.039125 (rank : 149) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MYO1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.043936 (rank : 138) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43795, O43794, Q7Z6L5 | Gene names | MYO1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin Ib (Myosin I alpha) (MMI-alpha) (MMIa) (MYH-1c). | |||||
|
MYO1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.044007 (rank : 137) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46735, P70244 | Gene names | Myo1b | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ib (Myosin I alpha) (MMI-alpha) (MMIa) (MIH-L). | |||||
|
CDT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.051735 (rank : 117) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
DGKD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.015898 (rank : 171) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
KPB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.011113 (rank : 177) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18826 | Gene names | Phka1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform (Phosphorylase kinase alpha M subunit). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.054089 (rank : 96) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
NOS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.021311 (rank : 164) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.020203 (rank : 167) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.057536 (rank : 79) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.019602 (rank : 168) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.043570 (rank : 140) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.042996 (rank : 142) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
CU013_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.037024 (rank : 150) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
K1219_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.020204 (rank : 166) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BQZ4, Q80TH5, Q8BQN1 | Gene names | Kiaa1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.024237 (rank : 161) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.015609 (rank : 173) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.022934 (rank : 162) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | -0.000453 (rank : 194) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.004357 (rank : 192) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CTNA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.029362 (rank : 157) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI47, Q5VSR2, Q6P056 | Gene names | CTNNA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Catenin alpha-3 (Alpha T-catenin) (Cadherin-associated protein). | |||||
|
GP128_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.005587 (rank : 187) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BM96, Q80T42 | Gene names | Gpr128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
LAMA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.032293 (rank : 155) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.055383 (rank : 82) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.054485 (rank : 91) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.053134 (rank : 107) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
USH2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.006223 (rank : 186) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
DOK5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.040756 (rank : 145) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P104, Q5T7Y0, Q5TE53, Q8TEW7, Q96H13, Q9BZ24, Q9NQF4, Q9Y411 | Gene names | DOK5, C20orf180 | |||
|
Domain Architecture |
|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (IRS6) (Protein dok-5). | |||||
|
DOK5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.040688 (rank : 147) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZM9, Q8BRI3, Q9CSM6 | Gene names | Dok5 | |||
|
Domain Architecture |
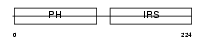
|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (Protein dok-5). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.015184 (rank : 174) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
FRMD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.269942 (rank : 50) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.059076 (rank : 75) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
MET7B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.043762 (rank : 139) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DD20, Q78ID2 | Gene names | Mettl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 7B precursor (EC 2.1.1.-). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.055366 (rank : 83) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.053245 (rank : 104) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.007741 (rank : 181) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.032678 (rank : 154) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
ABCD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.004220 (rank : 193) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61285, Q8BQ63, Q8C4B6 | Gene names | Abcd2, Aldr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family D member 2 (Adrenoleukodystrophy- related protein). | |||||
|
CA101_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.013138 (rank : 175) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SY80, Q8IYZ6 | Gene names | C1orf101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf101 precursor. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.004738 (rank : 190) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.042716 (rank : 143) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
K1949_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.010714 (rank : 178) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
KKCC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | -0.004253 (rank : 195) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5S9, Q9BQH3 | Gene names | CAMKK1, CAMKKA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
MGRN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.027824 (rank : 158) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D074, Q3U5V9, Q3UDA1, Q6ZQ97, Q8BZM9 | Gene names | Mgrn1, Kiaa0544 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.053657 (rank : 99) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
NPTX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.015753 (rank : 172) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |
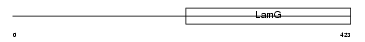
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.016258 (rank : 170) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |
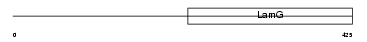
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
SIPA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.020798 (rank : 165) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.007374 (rank : 183) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.033629 (rank : 153) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TSC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.024653 (rank : 160) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.004530 (rank : 191) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.006455 (rank : 185) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
CCD91_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.053949 (rank : 97) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.047529 (rank : 134) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DGKZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.005046 (rank : 189) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13574, O00542 | Gene names | DGKZ, DAGK6 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase zeta (EC 2.7.1.107) (Diglyceride kinase zeta) (DGK-zeta) (DAG kinase zeta). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.040711 (rank : 146) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.008192 (rank : 180) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.052560 (rank : 113) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.059687 (rank : 74) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.062675 (rank : 70) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYO1E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.035044 (rank : 152) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.005314 (rank : 188) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.007700 (rank : 182) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
PO2F1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.006911 (rank : 184) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.012527 (rank : 176) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.036665 (rank : 151) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.055098 (rank : 87) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.052619 (rank : 112) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.067944 (rank : 68) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050020 (rank : 130) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.061550 (rank : 73) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.053930 (rank : 98) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.053270 (rank : 103) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.061976 (rank : 72) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.050986 (rank : 122) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.054321 (rank : 93) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054666 (rank : 89) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.050325 (rank : 128) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.053062 (rank : 108) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.051848 (rank : 116) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.053238 (rank : 105) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.053043 (rank : 109) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.053213 (rank : 106) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.050832 (rank : 124) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.054276 (rank : 95) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.055332 (rank : 85) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.054728 (rank : 88) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052745 (rank : 111) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050788 (rank : 125) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.055229 (rank : 86) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052338 (rank : 114) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.054497 (rank : 90) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.054476 (rank : 92) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NEMO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.054297 (rank : 94) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.064599 (rank : 69) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.050658 (rank : 126) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
REST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.055759 (rank : 81) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.051269 (rank : 121) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SAS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.050026 (rank : 129) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.051324 (rank : 120) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.051562 (rank : 119) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.062535 (rank : 71) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
TSNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.051864 (rank : 115) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.050879 (rank : 123) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
TLN2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 157 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
TLN2_MOUSE
|
||||||
| NC score | 0.996891 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.979773 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.978750 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
MESD1_MOUSE
|
||||||
| NC score | 0.574736 (rank : 5) | θ value | 4.9032e-22 (rank : 8) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERE8, Q542D0 | Gene names | Mesdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
MESD1_HUMAN
|
||||||
| NC score | 0.563966 (rank : 6) | θ value | 7.0802e-21 (rank : 9) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1K6 | Gene names | MESDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm development candidate 1. | |||||
|
E41LA_MOUSE
|
||||||
| NC score | 0.448232 (rank : 7) | θ value | 1.47974e-10 (rank : 13) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_HUMAN
|
||||||
| NC score | 0.441439 (rank : 8) | θ value | 9.59137e-10 (rank : 21) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41L5_HUMAN
|
||||||
| NC score | 0.437186 (rank : 9) | θ value | 1.9326e-10 (rank : 15) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.435811 (rank : 10) | θ value | 4.76016e-09 (rank : 31) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.434776 (rank : 11) | θ value | 4.76016e-09 (rank : 30) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.434680 (rank : 12) | θ value | 4.30538e-10 (rank : 18) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
E41LB_MOUSE
|
||||||
| NC score | 0.433099 (rank : 13) | θ value | 6.21693e-09 (rank : 32) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.417844 (rank : 14) | θ value | 9.59137e-10 (rank : 20) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.415217 (rank : 15) | θ value | 9.59137e-10 (rank : 19) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.412886 (rank : 16) | θ value | 5.26297e-08 (rank : 34) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.409333 (rank : 17) | θ value | 8.97725e-08 (rank : 36) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.408703 (rank : 18) | θ value | 1.17247e-07 (rank : 37) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.407490 (rank : 19) | θ value | 6.64225e-11 (rank : 10) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.406770 (rank : 20) | θ value | 6.64225e-11 (rank : 11) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.406328 (rank : 21) | θ value | 1.63604e-09 (rank : 26) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.405411 (rank : 22) | θ value | 2.79066e-09 (rank : 27) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FRMD5_MOUSE
|
||||||
| NC score | 0.404292 (rank : 23) | θ value | 7.1131e-05 (rank : 43) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.404119 (rank : 24) | θ value | 0.000121331 (rank : 45) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.399609 (rank : 25) | θ value | 5.81887e-07 (rank : 38) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.397518 (rank : 26) | θ value | 2.88788e-06 (rank : 40) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.384029 (rank : 27) | θ value | 1.25267e-09 (rank : 22) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.381100 (rank : 28) | θ value | 1.25267e-09 (rank : 23) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.377497 (rank : 29) | θ value | 0.000121331 (rank : 44) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.365201 (rank : 30) | θ value | 0.00020696 (rank : 47) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.364777 (rank : 31) | θ value | 0.00020696 (rank : 48) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.363703 (rank : 32) | θ value | 0.0113563 (rank : 53) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.361591 (rank : 33) | θ value | 0.000461057 (rank : 49) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.354471 (rank : 34) | θ value | 0.00134147 (rank : 51) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
PKHC1_HUMAN
|
||||||
| NC score | 0.345524 (rank : 35) | θ value | 2.52405e-10 (rank : 16) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
PKHC1_MOUSE
|
||||||
| NC score | 0.343303 (rank : 36) | θ value | 3.29651e-10 (rank : 17) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.341566 (rank : 37) | θ value | 0.0330416 (rank : 58) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
URP1_HUMAN
|
||||||
| NC score | 0.333293 (rank : 38) | θ value | 1.25267e-09 (rank : 25) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQL6, Q8IX34, Q8IYH2, Q9NWM2, Q9NXQ3 | Gene names | URP1, C20orf42, KIND1 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 1 (Kindlin-1) (Kindlerin) (Kindlin syndrome protein). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.331505 (rank : 39) | θ value | 1.99067e-23 (rank : 6) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
URP2_HUMAN
|
||||||
| NC score | 0.327734 (rank : 40) | θ value | 3.64472e-09 (rank : 28) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UX7, Q8IUA1, Q8N207, Q9BT48 | Gene names | URP2, KIND3, MIG2B | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3) (MIG2-like). | |||||
|
URP2_MOUSE
|
||||||
| NC score | 0.326038 (rank : 41) | θ value | 3.64472e-09 (rank : 29) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.326036 (rank : 42) | θ value | 0.000158464 (rank : 46) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
FRMD6_HUMAN
|
||||||
| NC score | 0.318498 (rank : 43) | θ value | 0.000602161 (rank : 50) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.307406 (rank : 44) | θ value | 1.25267e-09 (rank : 24) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.295600 (rank : 45) | θ value | 2.59989e-23 (rank : 7) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.294508 (rank : 46) | θ value | 2.36244e-08 (rank : 33) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.280127 (rank : 47) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.276201 (rank : 48) | θ value | 4.73814e-25 (rank : 5) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.271969 (rank : 49) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FRMD1_HUMAN
|
||||||
| NC score | 0.269942 (rank : 50) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.269036 (rank : 51) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.250974 (rank : 52) | θ value | 6.87365e-08 (rank : 35) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.250047 (rank : 53) | θ value | 0.00869519 (rank : 52) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.245496 (rank : 54) | θ value | 0.0148317 (rank : 55) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.242628 (rank : 55) | θ value | 0.0330416 (rank : 59) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.237869 (rank : 56) | θ value | 9.92553e-07 (rank : 39) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.237136 (rank : 57) | θ value | 0.47712 (rank : 74) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
KRIT1_HUMAN
|
||||||
| NC score | 0.202410 (rank : 58) | θ value | 3.77169e-06 (rank : 41) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |

|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
KRIT1_MOUSE
|
||||||
| NC score | 0.192427 (rank : 59) | θ value | 8.40245e-06 (rank : 42) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
MYO7A_MOUSE
|
||||||
| NC score | 0.133263 (rank : 60) | θ value | 1.47974e-10 (rank : 14) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97479 | Gene names | Myo7a, Myo7 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-7A (Myosin VIIa). | |||||
|
MYO7A_HUMAN
|
||||||
| NC score | 0.132745 (rank : 61) | θ value | 1.133e-10 (rank : 12) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13402, P78427, Q13321, Q14785, Q92821, Q92822 | Gene names | MYO7A, USH1B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-7A (Myosin VIIa). | |||||
|
VINC_HUMAN
|
||||||
| NC score | 0.120404 (rank : 62) | θ value | 0.0193708 (rank : 56) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18206, Q16450, Q5SWX2, Q7Z3B8, Q8IXU7 | Gene names | VCL | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
VINC_MOUSE
|
||||||
| NC score | 0.117034 (rank : 63) | θ value | 0.0193708 (rank : 57) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64727, Q8BP32, Q8BS46, Q922C5, Q922D9 | Gene names | Vcl | |||
|
Domain Architecture |

|
|||||
| Description | Vinculin (Metavinculin). | |||||
|
DOK4_HUMAN
|
||||||
| NC score | 0.095194 (rank : 64) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_MOUSE
|
||||||
| NC score | 0.095041 (rank : 65) | θ value | 0.0736092 (rank : 62) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
M6PBP_HUMAN
|
||||||
| NC score | 0.075910 (rank : 66) | θ value | 0.365318 (rank : 71) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60664, Q53G77, Q9BS03, Q9UBD7, Q9UP92 | Gene names | M6PRBP1, TIP47 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-6-phosphate receptor-binding protein 1 (Cargo selection protein TIP47) (47 kDa mannose 6-phosphate receptor-binding protein) (47 kDa MPR-binding protein) (Placental protein 17) (PP17). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.070101 (rank : 67) | θ value | 0.163984 (rank : 68) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.067944 (rank : 68) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.064599 (rank : 69) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.062675 (rank : 70) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.062535 (rank : 71) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.061976 (rank : 72) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.061550 (rank : 73) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.059687 (rank : 74) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.059076 (rank : 75) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
CTNA1_MOUSE
|
||||||
| NC score | 0.058891 (rank : 76) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26231 | Gene names | Ctnna1, Catna1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (102 kDa cadherin-associated protein) (CAP102) (Alpha E-catenin). | |||||
|
CTNA1_HUMAN
|
||||||
| NC score | 0.058857 (rank : 77) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35221, Q12795 | Gene names | CTNNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (Cadherin-associated protein) (Alpha E-catenin) (NY- REN-13 antigen). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.057780 (rank : 78) | θ value | 0.0961366 (rank : 63) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.057536 (rank : 79) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.057392 (rank : 80) | θ value | 0.163984 (rank : 67) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.055759 (rank : 81) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.055383 (rank : 82) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.055366 (rank : 83) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.055333 (rank : 84) | θ value | 0.125558 (rank : 64) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.055332 (rank : 85) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.055229 (rank : 86) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.055098 (rank : 87) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.054728 (rank : 88) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.054666 (rank : 89) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.054497 (rank : 90) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.054485 (rank : 91) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.054476 (rank : 92) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.054321 (rank : 93) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NEMO_MOUSE
|
||||||
| NC score | 0.054297 (rank : 94) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.054276 (rank : 95) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.054089 (rank : 96) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.053949 (rank : 97) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.053930 (rank : 98) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.053657 (rank : 99) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.053468 (rank : 100) | θ value | 0.125558 (rank : 65) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.053404 (rank : 101) | θ value | 0.125558 (rank : 66) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.053309 (rank : 102) | θ value | 0.813845 (rank : 76) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.053270 (rank : 103) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.053245 (rank : 104) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.053238 (rank : 105) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.053213 (rank : 106) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.053134 (rank : 107) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.053062 (rank : 108) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.053043 (rank : 109) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
DOK6_HUMAN
|
||||||
| NC score | 0.053009 (rank : 110) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PKX4, Q4V9S3, Q8WUZ8, Q96HI2, Q96LU2 | Gene names | DOK6, DOK5L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 6 (Downstream of tyrosine kinase 6). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.052745 (rank : 111) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.052619 (rank : 112) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.052560 (rank : 113) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.052338 (rank : 114) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
TSNA1_HUMAN
|
||||||
| NC score | 0.051864 (rank : 115) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.051848 (rank : 116) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
CDT1_HUMAN
|
||||||
| NC score | 0.051735 (rank : 117) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
CLC4F_MOUSE
|
||||||
| NC score | 0.051723 (rank : 118) | θ value | 0.0148317 (rank : 54) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.051562 (rank : 119) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.051324 (rank : 120) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.051269 (rank : 121) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.050986 (rank : 122) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.050879 (rank : 123) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.050832 (rank : 124) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.050788 (rank : 125) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.050658 (rank : 126) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.050456 (rank : 127) | θ value | 0.813845 (rank : 75) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.050325 (rank : 128) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.050026 (rank : 129) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.050020 (rank : 130) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.049986 (rank : 131) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.049393 (rank : 132) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.048752 (rank : 133) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.047529 (rank : 134) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.046744 (rank : 135) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.046311 (rank : 136) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
MYO1B_MOUSE
|
||||||
| NC score | 0.044007 (rank : 137) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46735, P70244 | Gene names | Myo1b | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ib (Myosin I alpha) (MMI-alpha) (MMIa) (MIH-L). | |||||
|
MYO1B_HUMAN
|
||||||
| NC score | 0.043936 (rank : 138) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43795, O43794, Q7Z6L5 | Gene names | MYO1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin Ib (Myosin I alpha) (MMI-alpha) (MMIa) (MYH-1c). | |||||
|
MET7B_MOUSE
|
||||||
| NC score | 0.043762 (rank : 139) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DD20, Q78ID2 | Gene names | Mettl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 7B precursor (EC 2.1.1.-). | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.043570 (rank : 140) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.043517 (rank : 141) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.042996 (rank : 142) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.042716 (rank : 143) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.041406 (rank : 144) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
DOK5_HUMAN
|
||||||
| NC score | 0.040756 (rank : 145) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P104, Q5T7Y0, Q5TE53, Q8TEW7, Q96H13, Q9BZ24, Q9NQF4, Q9Y411 | Gene names | DOK5, C20orf180 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (IRS6) (Protein dok-5). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.040711 (rank : 146) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
DOK5_MOUSE
|
||||||
| NC score | 0.040688 (rank : 147) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZM9, Q8BRI3, Q9CSM6 | Gene names | Dok5 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (Protein dok-5). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.039469 (rank : 148) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.039125 (rank : 149) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CU013_HUMAN
|
||||||
| NC score | 0.037024 (rank : 150) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.036665 (rank : 151) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
MYO1E_HUMAN
|
||||||
| NC score | 0.035044 (rank : 152) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.033629 (rank : 153) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.032678 (rank : 154) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
LAMA4_HUMAN
|
||||||
| NC score | 0.032293 (rank : 155) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.031064 (rank : 156) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
CTNA3_HUMAN
|
||||||
| NC score | 0.029362 (rank : 157) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI47, Q5VSR2, Q6P056 | Gene names | CTNNA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Catenin alpha-3 (Alpha T-catenin) (Cadherin-associated protein). | |||||
|
MGRN1_MOUSE
|
||||||
| NC score | 0.027824 (rank : 158) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D074, Q3U5V9, Q3UDA1, Q6ZQ97, Q8BZM9 | Gene names | Mgrn1, Kiaa0544 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.027498 (rank : 159) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
TSC2_HUMAN
|
||||||
| NC score | 0.024653 (rank : 160) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.024237 (rank : 161) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.022934 (rank : 162) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RNF6_HUMAN
|
||||||
| NC score | 0.022442 (rank : 163) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
NOS1_HUMAN
|
||||||
| NC score | 0.021311 (rank : 164) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.020798 (rank : 165) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
K1219_MOUSE
|
||||||
| NC score | 0.020204 (rank : 166) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BQZ4, Q80TH5, Q8BQN1 | Gene names | Kiaa1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.020203 (rank : 167) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.019602 (rank : 168) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
K1C16_HUMAN
|
||||||
| NC score | 0.018782 (rank : 169) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08779, P30654, Q16402, Q9UBG8 | Gene names | KRT16, KRT16A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
NPTX1_MOUSE
|
||||||
| NC score | 0.016258 (rank : 170) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
DGKD_HUMAN
|
||||||
| NC score | 0.015898 (rank : 171) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
NPTX1_HUMAN
|
||||||
| NC score | 0.015753 (rank : 172) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.015609 (rank : 173) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.015184 (rank : 174) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
CA101_HUMAN
|
||||||
| NC score | 0.013138 (rank : 175) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SY80, Q8IYZ6 | Gene names | C1orf101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf101 precursor. | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.012527 (rank : 176) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
KPB1_MOUSE
|
||||||
| NC score | 0.011113 (rank : 177) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18826 | Gene names | Phka1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform (Phosphorylase kinase alpha M subunit). | |||||
|
K1949_HUMAN
|
||||||
| NC score | 0.010714 (rank : 178) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
ITA4_HUMAN
|
||||||
| NC score | 0.010338 (rank : 179) | θ value | 1.38821 (rank : 89) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.008192 (rank : 180) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.007741 (rank : 181) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.007700 (rank : 182) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.007374 (rank : 183) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
PO2F1_MOUSE
|
||||||
| NC score | 0.006911 (rank : 184) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.006455 (rank : 185) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.006223 (rank : 186) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
GP128_MOUSE
|
||||||
| NC score | 0.005587 (rank : 187) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BM96, Q80T42 | Gene names | Gpr128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.005314 (rank : 188) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
DGKZ_HUMAN
|
||||||
| NC score | 0.005046 (rank : 189) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13574, O00542 | Gene names | DGKZ, DAGK6 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase zeta (EC 2.7.1.107) (Diglyceride kinase zeta) (DGK-zeta) (DAG kinase zeta). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.004738 (rank : 190) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.004530 (rank : 191) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.004357 (rank : 192) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
ABCD2_MOUSE
|
||||||
| NC score | 0.004220 (rank : 193) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61285, Q8BQ63, Q8C4B6 | Gene names | Abcd2, Aldr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family D member 2 (Adrenoleukodystrophy- related protein). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | -0.000453 (rank : 194) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
KKCC1_HUMAN
|
||||||
| NC score | -0.004253 (rank : 195) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5S9, Q9BQH3 | Gene names | CAMKK1, CAMKKA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||