Please be patient as the page loads
|
CA106_MOUSE
|
||||||
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CA106_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.965510 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 3) | NC score | 0.474466 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 4) | NC score | 0.475202 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 5) | NC score | 0.434925 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 6) | NC score | 0.428329 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 7) | NC score | 0.092594 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
THSD1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.102020 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.047736 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.070819 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.040870 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.077425 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.032919 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
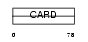
CARD6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.077512 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
CO039_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.058666 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.061072 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PP4R1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.054288 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.030428 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.028334 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
BACH1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.022871 (rank : 100) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.035695 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.036077 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.052198 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.036179 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.055561 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.048398 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.035650 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
ARRD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.048459 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.038308 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.068142 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.038998 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.048621 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.040943 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.069989 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CL025_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.041894 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.056825 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.014806 (rank : 113) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.052824 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.020347 (rank : 105) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.037413 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CDT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.037068 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.032274 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.036993 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.069948 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.011166 (rank : 117) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
KSYK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.005133 (rank : 127) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.017461 (rank : 109) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LYST_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.015557 (rank : 112) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.036451 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SIG10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.011211 (rank : 116) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.014536 (rank : 114) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
WDR67_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.029195 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NXY1, Q3TBW4, Q3U4F6, Q5KSA3, Q810J7, Q8C3Y2 | Gene names | Wdr67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67 (Protein 4-B-3). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.033098 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.060388 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.029753 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.013614 (rank : 115) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.007222 (rank : 124) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.009735 (rank : 118) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.019969 (rank : 106) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.018171 (rank : 108) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.004937 (rank : 128) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
APOE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.020735 (rank : 104) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08226 | Gene names | Apoe | |||
|
Domain Architecture |
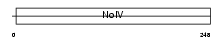
|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.051942 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FREA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.008625 (rank : 121) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
IRAK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.006939 (rank : 125) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43187 | Gene names | IRAK2 | |||
|
Domain Architecture |
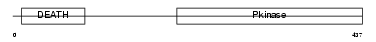
|
|||||
| Description | Interleukin-1 receptor-associated kinase-like 2 (IRAK-2). | |||||
|
KIF1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.005827 (rank : 126) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.034831 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.008128 (rank : 122) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.034612 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TMEPA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.017396 (rank : 110) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
ZN297_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.001930 (rank : 131) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15209, Q5STL0, Q5STR7, Q8WV82 | Gene names | ZNF297, BING1, ZBTB22A | |||
|
Domain Architecture |
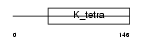
|
|||||
| Description | Zinc finger protein 297 (BING1 protein) (Zinc finger and BTB domain- containing protein 22A). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.003754 (rank : 130) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BT2A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.004637 (rank : 129) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7KYR7, O00475, P78408, Q7KYQ7, Q7Z386, Q9NU62 | Gene names | BTN2A1, BT2.1, BTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A1 precursor. | |||||
|
CJ078_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.021194 (rank : 102) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.025349 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.008776 (rank : 120) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.023246 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.022084 (rank : 101) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TJAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.020818 (rank : 103) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.008930 (rank : 119) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.035314 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.026781 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.060364 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.043423 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.040770 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.046979 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.017057 (rank : 111) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.019009 (rank : 107) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.007828 (rank : 123) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.080833 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.080531 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050217 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
E41L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.088825 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.088005 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.083585 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.081397 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.083605 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.082811 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.097785 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
E41LA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.095638 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.095450 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.091514 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
E41LB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.094084 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.105973 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.106303 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.081298 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.080877 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.073199 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FRMD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.065098 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.103454 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.102696 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.111280 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.100945 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
MERL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.111750 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.110931 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MOES_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.102306 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.100630 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.090115 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.094413 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051837 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058596 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PTN13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.054445 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.055668 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.067551 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.067954 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.067889 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.068986 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058856 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058423 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
RADI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.107133 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.106008 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.965510 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.475202 (rank : 3) | θ value | 3.74554e-30 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.474466 (rank : 4) | θ value | 9.8567e-31 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.434925 (rank : 5) | θ value | 4.58923e-20 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.428329 (rank : 6) | θ value | 2.5182e-18 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.111750 (rank : 7) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
FRMD6_HUMAN
|
||||||
| NC score | 0.111280 (rank : 8) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.110931 (rank : 9) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.107133 (rank : 10) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.106303 (rank : 11) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.106008 (rank : 12) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.105973 (rank : 13) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.103454 (rank : 14) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_MOUSE
|
||||||
| NC score | 0.102696 (rank : 15) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.102306 (rank : 16) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
THSD1_MOUSE
|
||||||
| NC score | 0.102020 (rank : 17) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.100945 (rank : 18) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.100630 (rank : 19) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
E41L5_HUMAN
|
||||||
| NC score | 0.097785 (rank : 20) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
E41LA_HUMAN
|
||||||
| NC score | 0.095638 (rank : 21) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| NC score | 0.095450 (rank : 22) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.094413 (rank : 23) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
E41LB_MOUSE
|
||||||
| NC score | 0.094084 (rank : 24) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.092594 (rank : 25) | θ value | 6.43352e-06 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.091514 (rank : 26) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.090115 (rank : 27) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.088825 (rank : 28) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.088005 (rank : 29) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.083605 (rank : 30) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.083585 (rank : 31) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.082811 (rank : 32) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.081397 (rank : 33) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.081298 (rank : 34) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.080877 (rank : 35) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.080833 (rank : 36) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.080531 (rank : 37) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
CARD6_HUMAN
|
||||||
| NC score | 0.077512 (rank : 38) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.077425 (rank : 39) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.073199 (rank : 40) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.070819 (rank : 41) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.069989 (rank : 42) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.069948 (rank : 43) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.068986 (rank : 44) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.068142 (rank : 45) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.067954 (rank : 46) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.067889 (rank : 47) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.067551 (rank : 48) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
FRMD1_HUMAN
|
||||||
| NC score | 0.065098 (rank : 49) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.061072 (rank : 50) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.060388 (rank : 51) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.060364 (rank : 52) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.058856 (rank : 53) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.058666 (rank : 54) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.058596 (rank : 55) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.058423 (rank : 56) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.056825 (rank : 57) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.055668 (rank : 58) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.055561 (rank : 59) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.054445 (rank : 60) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PP4R1_MOUSE
|
||||||
| NC score | 0.054288 (rank : 61) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.052824 (rank : 62) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.052198 (rank : 63) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.051942 (rank : 64) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.051837 (rank : 65) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.050217 (rank : 66) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.048621 (rank : 67) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.048459 (rank : 68) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.048398 (rank : 69) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.047736 (rank : 70) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.046979 (rank : 71) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.043423 (rank : 72) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CL025_HUMAN
|
||||||
| NC score | 0.041894 (rank : 73) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.040943 (rank : 74) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.040870 (rank : 75) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.040770 (rank : 76) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.038998 (rank : 77) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.038308 (rank : 78) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.037413 (rank : 79) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CDT1_HUMAN
|
||||||
| NC score | 0.037068 (rank : 80) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.036993 (rank : 81) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.036451 (rank : 82) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.036179 (rank : 83) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.036077 (rank : 84) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.035695 (rank : 85) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.035650 (rank : 86) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.035314 (rank : 87) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.034831 (rank : 88) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.034612 (rank : 89) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.033098 (rank : 90) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.032919 (rank : 91) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.032274 (rank : 92) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.030428 (rank : 93) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.029753 (rank : 94) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
WDR67_MOUSE
|
||||||
| NC score | 0.029195 (rank : 95) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NXY1, Q3TBW4, Q3U4F6, Q5KSA3, Q810J7, Q8C3Y2 | Gene names | Wdr67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67 (Protein 4-B-3). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.028334 (rank : 96) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.026781 (rank : 97) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.025349 (rank : 98) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.023246 (rank : 99) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
BACH1_MOUSE
|
||||||
| NC score | 0.022871 (rank : 100) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.022084 (rank : 101) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
CJ078_HUMAN
|
||||||
| NC score | 0.021194 (rank : 102) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
TJAP1_HUMAN
|
||||||
| NC score | 0.020818 (rank : 103) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
APOE_MOUSE
|
||||||
| NC score | 0.020735 (rank : 104) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08226 | Gene names | Apoe | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.020347 (rank : 105) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.019969 (rank : 106) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.019009 (rank : 107) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.018171 (rank : 108) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.017461 (rank : 109) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
TMEPA_MOUSE
|
||||||
| NC score | 0.017396 (rank : 110) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.017057 (rank : 111) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.015557 (rank : 112) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.014806 (rank : 113) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.014536 (rank : 114) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.013614 (rank : 115) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
SIG10_HUMAN
|
||||||
| NC score | 0.011211 (rank : 116) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.011166 (rank : 117) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.009735 (rank : 118) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.008930 (rank : 119) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.008776 (rank : 120) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
FREA_HUMAN
|
||||||
| NC score | 0.008625 (rank : 121) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.008128 (rank : 122) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.007828 (rank : 123) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.007222 (rank : 124) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
IRAK2_HUMAN
|
||||||
| NC score | 0.006939 (rank : 125) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43187 | Gene names | IRAK2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase-like 2 (IRAK-2). | |||||
|
KIF1C_HUMAN
|
||||||
| NC score | 0.005827 (rank : 126) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
KSYK_MOUSE
|
||||||
| NC score | 0.005133 (rank : 127) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.004937 (rank : 128) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
BT2A1_HUMAN
|
||||||
| NC score | 0.004637 (rank : 129) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7KYR7, O00475, P78408, Q7KYQ7, Q7Z386, Q9NU62 | Gene names | BTN2A1, BT2.1, BTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A1 precursor. | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.003754 (rank : 130) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ZN297_HUMAN
|
||||||
| NC score | 0.001930 (rank : 131) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15209, Q5STL0, Q5STR7, Q8WV82 | Gene names | ZNF297, BING1, ZBTB22A | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 297 (BING1 protein) (Zinc finger and BTB domain- containing protein 22A). | |||||