Please be patient as the page loads
|
KCNH2_HUMAN
|
||||||
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNH2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995949 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.979253 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.988809 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.989521 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 4.75757e-126 (rank : 6) | NC score | 0.960370 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 3.19119e-122 (rank : 7) | NC score | 0.954467 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 7.10925e-122 (rank : 8) | NC score | 0.954225 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 1.21265e-121 (rank : 9) | NC score | 0.953508 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 1.58377e-121 (rank : 10) | NC score | 0.953719 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 6.01832e-121 (rank : 11) | NC score | 0.957521 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 2.29227e-112 (rank : 12) | NC score | 0.951447 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 2.9938e-112 (rank : 13) | NC score | 0.951879 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 14) | NC score | 0.792173 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 15) | NC score | 0.788487 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 1.28434e-38 (rank : 16) | NC score | 0.805448 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 17) | NC score | 0.795386 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 18) | NC score | 0.790299 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 19) | NC score | 0.808720 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 20) | NC score | 0.804862 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 21) | NC score | 0.805358 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 22) | NC score | 0.808740 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 3.86705e-35 (rank : 23) | NC score | 0.822833 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 24) | NC score | 0.811748 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 25) | NC score | 0.812051 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 26) | NC score | 0.807246 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 27) | NC score | 0.754483 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 28) | NC score | 0.739519 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 29) | NC score | 0.732789 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 30) | NC score | 0.247086 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 31) | NC score | 0.184076 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 32) | NC score | 0.183439 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.188406 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.018011 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.009483 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CA106_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 36) | NC score | 0.032919 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 37) | NC score | 0.035228 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 38) | NC score | 0.174277 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 39) | NC score | 0.040324 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CP007_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 40) | NC score | 0.030492 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 41) | NC score | 0.066273 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 42) | NC score | 0.175650 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 43) | NC score | 0.173704 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.171969 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.028925 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 46) | NC score | 0.043157 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
CEBPE_MOUSE
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.025920 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.044526 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.044627 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
FRS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.026018 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.037579 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.058781 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.058770 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.019791 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.036713 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.024936 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.060326 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.163786 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.152788 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.010187 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.031570 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.144987 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.044174 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
NRG2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.024838 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.032224 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.032217 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.156355 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.146953 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.147096 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.101172 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.041089 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.006144 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
FRS3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.022513 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
GP124_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.007452 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
LYSM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.021117 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IV50, Q5CZ88, Q8WTV3 | Gene names | LYSMD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 2. | |||||
|
NRG2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.025312 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56974 | Gene names | Nrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Divergent of neuregulin 1) (DON-1)]. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.009421 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.160715 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
PKP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.008671 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.027393 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.017259 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.124842 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.031591 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MNAB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.013768 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.013620 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
KAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.131878 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.129577 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.055458 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.055471 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.123855 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.004967 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.020482 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
RGS14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.014110 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.029905 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.024612 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ATPD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.027400 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30049 | Gene names | ATP5D | |||
|
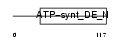
Domain Architecture |
|
|||||
| Description | ATP synthase delta chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.012646 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.059908 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.060016 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.120183 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.040954 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.040031 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.055277 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.004243 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.006568 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
TSSC4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.014807 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5U2, Q86VL2, Q9BRS6 | Gene names | TSSC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4 (Tumor-suppressing subchromosomal transferable fragment candidate gene 4 protein) (Tumor-suppressing STF cDNA 4 protein). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | -0.003559 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ZP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.014278 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62005, Q62016 | Gene names | Zp1 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
ANR25_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.004583 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
CQ10A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.011010 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MF6, Q6GMR6, Q6UWB9, Q86X16, Q96MF1, Q9BUP4 | Gene names | COQ10A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein COQ10 A, mitochondrial precursor. | |||||
|
HXB5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | -0.000423 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09067, P09069 | Gene names | HOXB5, HOX2A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B5 (Hox-2A) (HHO.C10) (HU-1). | |||||
|
KCNKC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.078281 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.001724 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.005168 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.027363 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.012349 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.039529 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.043619 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
XYLT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.007499 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
3BHS7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.011101 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQC1 | Gene names | Hsd3b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 7 (3 beta-hydroxysteroid dehydrogenase type VII) (3Beta-HSD VII) (3-beta-hydroxy-delta(5)-C27 steroid oxidoreductase) (EC 1.1.1.-) (C(27) 3beta-HSD). | |||||
|
ATG9A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.011296 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
CN004_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.013326 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
DCAK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | -0.002051 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PGN3, Q8BUU0, Q8BX25 | Gene names | Dcamkl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL2 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 2). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.014944 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.080348 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.034334 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
M3K10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | -0.002484 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
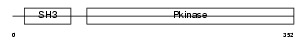
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.001896 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.032577 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.026803 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CI150_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.019266 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2P1, Q9D150 | Gene names | D4Bwg0951e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf150 homolog. | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | -0.000953 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.010324 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.030975 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.053144 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.004601 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
REL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.015642 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47932 | Gene names | Rln1, Rln, Rlx | |||
|
Domain Architecture |

|
|||||
| Description | Prorelaxin 1 precursor [Contains: Relaxin B chain; Relaxin A chain]. | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.022432 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TASP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.007546 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1G1, Q99JP3 | Gene names | Tasp1 | |||
|
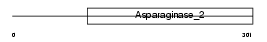
Domain Architecture |
|
|||||
| Description | Threonine aspartase 1 (EC 3.4.25.-) (Taspase-1) [Contains: Threonine aspartase subunit alpha; Threonine aspartase subunit beta]. | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.003637 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
TGIF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.004251 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15583, Q6ICR0, Q8N5X9, Q9NRS0 | Gene names | TGIF | |||
|
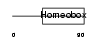
Domain Architecture |
|
|||||
| Description | 5'-TG-3'-interacting factor (Homeobox protein TGIF). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.015506 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
ZNF31_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | -0.003036 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17040, Q96H84 | Gene names | ZNF31, KOX29, ZNF360, ZSCAN20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 31 (Zinc finger protein KOX29) (Zinc finger and SCAN domain-containing protein 20) (Zinc finger protein 360). | |||||
|
ABD12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.011697 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2K0, Q5T710, Q96CR1, Q9BX05, Q9NPX7, Q9UFV6 | Gene names | Abhd12, C20orf22 | |||
|
Domain Architecture |

|
|||||
| Description | Abhydrolase domain-containing protein 12. | |||||
|
ABD12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.011691 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LR1, Q99M06 | Gene names | Abhd12 | |||
|
Domain Architecture |

|
|||||
| Description | Abhydrolase domain-containing protein 12. | |||||
|
AF17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.011179 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.014401 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CC054_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.007833 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96EL1 | Gene names | C3orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf54. | |||||
|
CN004_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.011628 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.003270 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | -0.000957 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
PITM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.003002 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
SOCS7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.007756 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.004758 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.003141 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.004475 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TOM40_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.004862 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96008, Q86VW4, Q8WY09, Q8WY10, Q8WY11, Q9BR95 | Gene names | TOMM40, PEREC1, TOM40 | |||
|
Domain Architecture |
|
|||||
| Description | Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) (Haymaker protein) (p38.5). | |||||
|
VASP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.015468 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
CT152_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.068259 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.090259 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.092409 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.089474 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.092842 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KCNK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.053513 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.070812 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.995949 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.989521 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.988809 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.979253 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.960370 (rank : 6) | θ value | 4.75757e-126 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.957521 (rank : 7) | θ value | 6.01832e-121 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.954467 (rank : 8) | θ value | 3.19119e-122 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.954225 (rank : 9) | θ value | 7.10925e-122 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.953719 (rank : 10) | θ value | 1.58377e-121 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.953508 (rank : 11) | θ value | 1.21265e-121 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.951879 (rank : 12) | θ value | 2.9938e-112 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.951447 (rank : 13) | θ value | 2.29227e-112 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.822833 (rank : 14) | θ value | 3.86705e-35 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.812051 (rank : 15) | θ value | 5.584e-34 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.811748 (rank : 16) | θ value | 1.12514e-34 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.808740 (rank : 17) | θ value | 2.67802e-36 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.808720 (rank : 18) | θ value | 2.86122e-38 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.807246 (rank : 19) | θ value | 2.12192e-33 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.805448 (rank : 20) | θ value | 1.28434e-38 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.805358 (rank : 21) | θ value | 4.88048e-38 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.804862 (rank : 22) | θ value | 2.86122e-38 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.795386 (rank : 23) | θ value | 2.19075e-38 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.792173 (rank : 24) | θ value | 1.98146e-39 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.790299 (rank : 25) | θ value | 2.19075e-38 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.788487 (rank : 26) | θ value | 9.83387e-39 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.754483 (rank : 27) | θ value | 2.87452e-22 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.739519 (rank : 28) | θ value | 2.27762e-19 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.732789 (rank : 29) | θ value | 5.60996e-18 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.247086 (rank : 30) | θ value | 0.00298849 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.188406 (rank : 31) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.184076 (rank : 32) | θ value | 0.0113563 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.183439 (rank : 33) | θ value | 0.0113563 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.175650 (rank : 34) | θ value | 0.0961366 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.174277 (rank : 35) | θ value | 0.0736092 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.173704 (rank : 36) | θ value | 0.0961366 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.171969 (rank : 37) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.163786 (rank : 38) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.160715 (rank : 39) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.156355 (rank : 40) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.152788 (rank : 41) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.147096 (rank : 42) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.146953 (rank : 43) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.144987 (rank : 44) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.131878 (rank : 45) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.129577 (rank : 46) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.124842 (rank : 47) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.123855 (rank : 48) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.120183 (rank : 49) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.101172 (rank : 50) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.092842 (rank : 51) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.092409 (rank : 52) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.090259 (rank : 53) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.089474 (rank : 54) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.080348 (rank : 55) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKC_HUMAN
|
||||||
| NC score | 0.078281 (rank : 56) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.070812 (rank : 57) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.068259 (rank : 58) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.066273 (rank : 59) | θ value | 0.0961366 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.060326 (rank : 60) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.060016 (rank : 61) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.059908 (rank : 62) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.058781 (rank : 63) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.058770 (rank : 64) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.055471 (rank : 65) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.055458 (rank : 66) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.055277 (rank : 67) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNK7_HUMAN
|
||||||
| NC score | 0.053513 (rank : 68) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.053144 (rank : 69) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.044627 (rank : 70) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.044526 (rank : 71) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.044174 (rank : 72) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.043619 (rank : 73) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.043157 (rank : 74) | θ value | 0.163984 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.041089 (rank : 75) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.040954 (rank : 76) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.040324 (rank : 77) | θ value | 0.0736092 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.040031 (rank : 78) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.039529 (rank : 79) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.037579 (rank : 80) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.036713 (rank : 81) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.035228 (rank : 82) | θ value | 0.0736092 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.034334 (rank : 83) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.032919 (rank : 84) | θ value | 0.0736092 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.032577 (rank : 85) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.032224 (rank : 86) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.032217 (rank : 87) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.031591 (rank : 88) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.031570 (rank : 89) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.030975 (rank : 90) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.030492 (rank : 91) | θ value | 0.0961366 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.029905 (rank : 92) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.028925 (rank : 93) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
ATPD_HUMAN
|
||||||
| NC score | 0.027400 (rank : 94) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30049 | Gene names | ATP5D | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase delta chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.027393 (rank : 95) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.027363 (rank : 96) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.026803 (rank : 97) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
FRS3_MOUSE
|
||||||
| NC score | 0.026018 (rank : 98) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
CEBPE_MOUSE
|
||||||
| NC score | 0.025920 (rank : 99) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
NRG2_MOUSE
|
||||||
| NC score | 0.025312 (rank : 100) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56974 | Gene names | Nrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Divergent of neuregulin 1) (DON-1)]. | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.024936 (rank : 101) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
NRG2_HUMAN
|
||||||
| NC score | 0.024838 (rank : 102) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.024612 (rank : 103) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
FRS3_HUMAN
|
||||||
| NC score | 0.022513 (rank : 104) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.022432 (rank : 105) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
LYSM2_HUMAN
|
||||||
| NC score | 0.021117 (rank : 106) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IV50, Q5CZ88, Q8WTV3 | Gene names | LYSMD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 2. | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.020482 (rank : 107) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.019791 (rank : 108) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CI150_MOUSE
|
||||||
| NC score | 0.019266 (rank : 109) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2P1, Q9D150 | Gene names | D4Bwg0951e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf150 homolog. | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.018011 (rank : 110) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.017259 (rank : 111) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
REL1_MOUSE
|
||||||
| NC score | 0.015642 (rank : 112) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47932 | Gene names | Rln1, Rln, Rlx | |||
|
Domain Architecture |

|
|||||
| Description | Prorelaxin 1 precursor [Contains: Relaxin B chain; Relaxin A chain]. | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.015506 (rank : 113) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.015468 (rank : 114) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.014944 (rank : 115) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
TSSC4_HUMAN
|
||||||
| NC score | 0.014807 (rank : 116) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5U2, Q86VL2, Q9BRS6 | Gene names | TSSC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4 (Tumor-suppressing subchromosomal transferable fragment candidate gene 4 protein) (Tumor-suppressing STF cDNA 4 protein). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.014401 (rank : 117) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ZP1_MOUSE
|
||||||
| NC score | 0.014278 (rank : 118) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62005, Q62016 | Gene names | Zp1 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.014110 (rank : 119) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
MNAB_HUMAN
|
||||||
| NC score | 0.013768 (rank : 120) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.013620 (rank : 121) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.013326 (rank : 122) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.012646 (rank : 123) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.012349 (rank : 124) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
ABD12_HUMAN
|
||||||
| NC score | 0.011697 (rank : 125) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2K0, Q5T710, Q96CR1, Q9BX05, Q9NPX7, Q9UFV6 | Gene names | Abhd12, C20orf22 | |||
|
Domain Architecture |

|
|||||
| Description | Abhydrolase domain-containing protein 12. | |||||
|
ABD12_MOUSE
|
||||||
| NC score | 0.011691 (rank : 126) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LR1, Q99M06 | Gene names | Abhd12 | |||
|
Domain Architecture |

|
|||||
| Description | Abhydrolase domain-containing protein 12. | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.011628 (rank : 127) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
ATG9A_HUMAN
|
||||||
| NC score | 0.011296 (rank : 128) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.011179 (rank : 129) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
3BHS7_MOUSE
|
||||||
| NC score | 0.011101 (rank : 130) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQC1 | Gene names | Hsd3b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 7 (3 beta-hydroxysteroid dehydrogenase type VII) (3Beta-HSD VII) (3-beta-hydroxy-delta(5)-C27 steroid oxidoreductase) (EC 1.1.1.-) (C(27) 3beta-HSD). | |||||
|
CQ10A_HUMAN
|
||||||
| NC score | 0.011010 (rank : 131) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MF6, Q6GMR6, Q6UWB9, Q86X16, Q96MF1, Q9BUP4 | Gene names | COQ10A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein COQ10 A, mitochondrial precursor. | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.010324 (rank : 132) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.010187 (rank : 133) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.009483 (rank : 134) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.009421 (rank : 135) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.008671 (rank : 136) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
CC054_HUMAN
|
||||||
| NC score | 0.007833 (rank : 137) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96EL1 | Gene names | C3orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf54. | |||||
|
SOCS7_MOUSE
|
||||||
| NC score | 0.007756 (rank : 138) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
TASP1_MOUSE
|
||||||
| NC score | 0.007546 (rank : 139) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1G1, Q99JP3 | Gene names | Tasp1 | |||
|
Domain Architecture |

|
|||||
| Description | Threonine aspartase 1 (EC 3.4.25.-) (Taspase-1) [Contains: Threonine aspartase subunit alpha; Threonine aspartase subunit beta]. | |||||
|
XYLT1_HUMAN
|
||||||
| NC score | 0.007499 (rank : 140) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.007452 (rank : 141) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.006568 (rank : 142) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.006144 (rank : 143) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.005168 (rank : 144) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.004967 (rank : 145) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
TOM40_HUMAN
|
||||||
| NC score | 0.004862 (rank : 146) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96008, Q86VW4, Q8WY09, Q8WY10, Q8WY11, Q9BR95 | Gene names | TOMM40, PEREC1, TOM40 | |||
|
Domain Architecture |

|
|||||
| Description | Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) (Haymaker protein) (p38.5). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.004758 (rank : 147) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.004601 (rank : 148) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
ANR25_HUMAN
|
||||||
| NC score | 0.004583 (rank : 149) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.004475 (rank : 150) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TGIF_HUMAN
|
||||||
| NC score | 0.004251 (rank : 151) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15583, Q6ICR0, Q8N5X9, Q9NRS0 | Gene names | TGIF | |||
|
Domain Architecture |

|
|||||
| Description | 5'-TG-3'-interacting factor (Homeobox protein TGIF). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.004243 (rank : 152) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.003637 (rank : 153) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.003270 (rank : 154) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.003141 (rank : 155) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
PITM1_MOUSE
|
||||||
| NC score | 0.003002 (rank : 156) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.001896 (rank : 157) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.001724 (rank : 158) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
HXB5_HUMAN
|
||||||
| NC score | -0.000423 (rank : 159) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09067, P09069 | Gene names | HOXB5, HOX2A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B5 (Hox-2A) (HHO.C10) (HU-1). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | -0.000953 (rank : 160) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | -0.000957 (rank : 161) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
DCAK2_MOUSE
|
||||||
| NC score | -0.002051 (rank : 162) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PGN3, Q8BUU0, Q8BX25 | Gene names | Dcamkl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL2 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 2). | |||||
|
M3K10_HUMAN
|
||||||
| NC score | -0.002484 (rank : 163) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
ZNF31_HUMAN
|
||||||
| NC score | -0.003036 (rank : 164) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17040, Q96H84 | Gene names | ZNF31, KOX29, ZNF360, ZSCAN20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 31 (Zinc finger protein KOX29) (Zinc finger and SCAN domain-containing protein 20) (Zinc finger protein 360). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.003559 (rank : 165) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||