Please be patient as the page loads
|
CT152_MOUSE
|
||||||
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CT152_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.980696 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
CT152_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 3) | NC score | 0.134084 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 4) | NC score | 0.134725 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
RPGF3_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 5) | NC score | 0.385360 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF3_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 6) | NC score | 0.387599 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 7) | NC score | 0.132826 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 8) | NC score | 0.132721 (rank : 39) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KGP2_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 9) | NC score | 0.132894 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 10) | NC score | 0.352593 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 11) | NC score | 0.354364 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
KAP2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 12) | NC score | 0.436976 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 13) | NC score | 0.449409 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 14) | NC score | 0.444008 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 15) | NC score | 0.442476 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 16) | NC score | 0.444732 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 17) | NC score | 0.426030 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.207994 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KAP3_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 19) | NC score | 0.394472 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 20) | NC score | 0.390826 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 21) | NC score | 0.174437 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.161990 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.164023 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.182681 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.180402 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.147811 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.148070 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.155294 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.153239 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.153142 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.147806 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.146894 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.157536 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.159430 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.024406 (rank : 60) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.151174 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.149528 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
MRP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.004792 (rank : 61) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15438, O60265, O60922, O75621, O95078, O95289, O95290, Q9UN52 | Gene names | ABCC3, CMOAT2, MLP2, MRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 2 (ATP-binding cassette sub-family C member 3) (Multidrug resistance-associated protein 3) (Multi-specific organic anion transporter-D) (MOAT-D). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.056553 (rank : 55) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.055306 (rank : 56) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
GRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.066099 (rank : 48) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.067146 (rank : 47) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.067183 (rank : 46) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.068259 (rank : 44) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.068160 (rank : 45) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.059874 (rank : 53) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.061134 (rank : 50) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.059457 (rank : 54) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.060447 (rank : 52) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.061683 (rank : 49) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.072768 (rank : 41) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.071043 (rank : 42) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.070392 (rank : 43) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.060555 (rank : 51) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.096529 (rank : 40) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.138911 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
RPGF5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.160892 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92565, Q8IXU5 | Gene names | RAPGEF5, GFR, KIAA0277, MRGEF | |||
|
Domain Architecture |
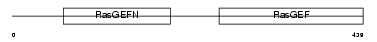
|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (Related to Epac) (Repac) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
RPGF5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.163377 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0Q9, Q8BJJ9, Q8C0R5 | Gene names | Rapgef5, Gfr, Kiaa0277, Mrgef | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.053930 (rank : 58) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.053633 (rank : 59) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SOS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.055060 (rank : 57) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
CT152_HUMAN
|
||||||
| NC score | 0.980696 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.449409 (rank : 3) | θ value | 4.1701e-05 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.444732 (rank : 4) | θ value | 0.000158464 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.444008 (rank : 5) | θ value | 0.000121331 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.442476 (rank : 6) | θ value | 0.000121331 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.436976 (rank : 7) | θ value | 2.44474e-05 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.426030 (rank : 8) | θ value | 0.000270298 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.394472 (rank : 9) | θ value | 0.00390308 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.390826 (rank : 10) | θ value | 0.0113563 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
RPGF3_MOUSE
|
||||||
| NC score | 0.387599 (rank : 11) | θ value | 1.38499e-08 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
RPGF3_HUMAN
|
||||||
| NC score | 0.385360 (rank : 12) | θ value | 8.11959e-09 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.354364 (rank : 13) | θ value | 2.21117e-06 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.352593 (rank : 14) | θ value | 1.69304e-06 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.207994 (rank : 15) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.182681 (rank : 16) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.180402 (rank : 17) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.174437 (rank : 18) | θ value | 0.0148317 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.164023 (rank : 19) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
RPGF5_MOUSE
|
||||||
| NC score | 0.163377 (rank : 20) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0Q9, Q8BJJ9, Q8C0R5 | Gene names | Rapgef5, Gfr, Kiaa0277, Mrgef | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.161990 (rank : 21) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
RPGF5_HUMAN
|
||||||
| NC score | 0.160892 (rank : 22) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92565, Q8IXU5 | Gene names | RAPGEF5, GFR, KIAA0277, MRGEF | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 5 (Guanine nucleotide exchange factor for Rap1) (Related to Epac) (Repac) (M-Ras-regulated Rap GEF) (MR-GEF). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.159430 (rank : 23) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.157536 (rank : 24) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.155294 (rank : 25) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.153239 (rank : 26) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.153142 (rank : 27) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.151174 (rank : 28) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.149528 (rank : 29) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.148070 (rank : 30) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.147811 (rank : 31) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.147806 (rank : 32) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.146894 (rank : 33) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.138911 (rank : 34) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.134725 (rank : 35) | θ value | 4.76016e-09 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.134084 (rank : 36) | θ value | 4.76016e-09 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP2_MOUSE
|
||||||
| NC score | 0.132894 (rank : 37) | θ value | 1.99992e-07 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.132826 (rank : 38) | θ value | 5.26297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.132721 (rank : 39) | θ value | 1.17247e-07 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.096529 (rank : 40) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.072768 (rank : 41) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.071043 (rank : 42) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.070392 (rank : 43) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.068259 (rank : 44) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.068160 (rank : 45) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.067183 (rank : 46) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.067146 (rank : 47) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
GRP3_HUMAN
|
||||||
| NC score | 0.066099 (rank : 48) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.061683 (rank : 49) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.061134 (rank : 50) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.060555 (rank : 51) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.060447 (rank : 52) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.059874 (rank : 53) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.059457 (rank : 54) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.056553 (rank : 55) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.055306 (rank : 56) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.055060 (rank : 57) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.053930 (rank : 58) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.053633 (rank : 59) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.024406 (rank : 60) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
MRP3_HUMAN
|
||||||
| NC score | 0.004792 (rank : 61) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15438, O60265, O60922, O75621, O95078, O95289, O95290, Q9UN52 | Gene names | ABCC3, CMOAT2, MLP2, MRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 2 (ATP-binding cassette sub-family C member 3) (Multidrug resistance-associated protein 3) (Multi-specific organic anion transporter-D) (MOAT-D). | |||||