Please be patient as the page loads
|
HCN1_HUMAN
|
||||||
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HCN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990656 (rank : 2) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.964151 (rank : 8) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.974583 (rank : 5) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.988166 (rank : 4) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.988545 (rank : 3) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.973135 (rank : 6) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.970029 (rank : 7) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 9) | NC score | 0.868271 (rank : 9) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 2.04573e-44 (rank : 10) | NC score | 0.856005 (rank : 12) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 11) | NC score | 0.857454 (rank : 11) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | 8.59492e-43 (rank : 12) | NC score | 0.861301 (rank : 10) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 13) | NC score | 0.854856 (rank : 13) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 14) | NC score | 0.812057 (rank : 17) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 15) | NC score | 0.808160 (rank : 19) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 16) | NC score | 0.808720 (rank : 18) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 17) | NC score | 0.807813 (rank : 20) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 18) | NC score | 0.798895 (rank : 23) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 19) | NC score | 0.799174 (rank : 22) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 20) | NC score | 0.802843 (rank : 21) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 21) | NC score | 0.792658 (rank : 27) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 8.61488e-35 (rank : 22) | NC score | 0.792858 (rank : 26) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 23) | NC score | 0.829583 (rank : 15) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 24) | NC score | 0.798329 (rank : 24) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 6.82597e-32 (rank : 25) | NC score | 0.793221 (rank : 25) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 26) | NC score | 0.836323 (rank : 14) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 27) | NC score | 0.820264 (rank : 16) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 28) | NC score | 0.781330 (rank : 28) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 29) | NC score | 0.781208 (rank : 29) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 30) | NC score | 0.301563 (rank : 30) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KGP2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 31) | NC score | 0.072640 (rank : 44) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.072576 (rank : 45) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KAP3_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 33) | NC score | 0.233340 (rank : 37) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 34) | NC score | 0.238765 (rank : 36) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 35) | NC score | 0.035332 (rank : 71) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 36) | NC score | 0.073335 (rank : 43) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 37) | NC score | 0.073587 (rank : 42) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KAP2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 38) | NC score | 0.242676 (rank : 35) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 39) | NC score | 0.246775 (rank : 34) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 40) | NC score | 0.073591 (rank : 41) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 41) | NC score | 0.043498 (rank : 62) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
KAP1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 42) | NC score | 0.264236 (rank : 32) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 43) | NC score | 0.261150 (rank : 33) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
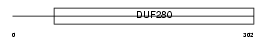
CCR3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.016526 (rank : 99) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51678 | Gene names | Ccr3, Cmkbr1l2, Cmkbr3 | |||
|
Domain Architecture |
|
|||||
| Description | Probable C-C chemokine receptor type 3 (C-C CKR-3) (CC-CKR-3) (CCR-3) (CCR3) (CKR3) (Macrophage inflammatory protein 1-alpha receptor-like 2) (MIP-1 alpha RL2) (CD193 antigen). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 45) | NC score | 0.271079 (rank : 31) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 46) | NC score | 0.014898 (rank : 102) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
DCP1B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.042134 (rank : 64) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.041390 (rank : 65) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.035371 (rank : 70) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.021725 (rank : 89) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.027030 (rank : 80) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.017498 (rank : 96) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.009452 (rank : 112) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.016928 (rank : 98) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.043069 (rank : 63) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
PC11X_HUMAN
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.002206 (rank : 130) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.028283 (rank : 78) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.022275 (rank : 87) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.014379 (rank : 104) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.034941 (rank : 72) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
PRGC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.021331 (rank : 90) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
SIG10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.006173 (rank : 122) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.022766 (rank : 86) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.025395 (rank : 81) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.017732 (rank : 95) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.047089 (rank : 61) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.018458 (rank : 94) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
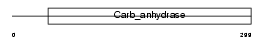
CAH11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.009948 (rank : 111) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75493, O60596, Q9UEC4 | Gene names | CA11, CARP2 | |||
|
Domain Architecture |
|
|||||
| Description | Carbonic anhydrase-related protein 2 precursor (CARP-2) (CA-RP II) (CA-XI) (Carbonic anhydrase-related protein 11) (CARP XI) (CA-RP XI). | |||||
|
CT152_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.147806 (rank : 38) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.038194 (rank : 68) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.038492 (rank : 67) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.050173 (rank : 60) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.015636 (rank : 101) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
PP14A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.039423 (rank : 66) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VC7, Q99MB9 | Gene names | Ppp1r14a, Cpi17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14A (17 kDa PKC-potentiated inhibitory protein of PP1) (CPI-17). | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.003543 (rank : 127) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
HRSL5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.010568 (rank : 110) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPX5, Q9CVQ2, Q9CVS0 | Gene names | Hrasls5, Hrlp5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
MYPN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.004871 (rank : 124) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
SCYL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.012864 (rank : 107) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
ZN707_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | -0.004003 (rank : 134) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96C28 | Gene names | ZNF707 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 707. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.014564 (rank : 103) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CAH11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.008729 (rank : 114) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70354, Q8VEA9 | Gene names | Ca11, Car11, Carp2 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase-related protein 2 precursor (CARP-2) (CA-RP II) (CA-XI) (Carbonic anhydrase-related protein 11) (CARP XI) (CA-RP XI). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.016491 (rank : 100) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
MAML1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.028517 (rank : 77) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.031375 (rank : 73) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.030254 (rank : 75) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.008407 (rank : 117) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PRPF3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.011373 (rank : 109) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
RL4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.012912 (rank : 106) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36578, P39029, Q4VBR0, Q969Z9 | Gene names | RPL4, RPL1 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L4 (L1). | |||||
|
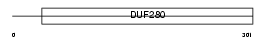
TAAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.002737 (rank : 129) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RJ0, Q3MIH8, Q5VUQ1 | Gene names | TAAR1, TA1, TAR1, TRAR1 | |||
|
Domain Architecture |
|
|||||
| Description | Trace amine-associated receptor 1 (Trace amine receptor 1) (TaR-1). | |||||
|
ANR41_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.003779 (rank : 126) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
GCP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.008442 (rank : 116) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.009413 (rank : 113) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.020931 (rank : 91) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.051719 (rank : 58) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.023079 (rank : 85) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.000150 (rank : 132) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.029397 (rank : 76) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.022060 (rank : 88) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
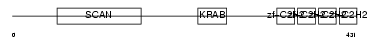
ZN500_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | -0.004326 (rank : 135) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 500. | |||||
|
CD248_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.002805 (rank : 128) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.025272 (rank : 82) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
ELL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.006697 (rank : 121) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
HMX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | -0.000792 (rank : 133) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42581 | Gene names | Hmx3, Nkx-5.1, Nkx5-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HMX3 (Homeobox protein Nkx-5.1). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.013469 (rank : 105) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.027998 (rank : 79) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.017097 (rank : 97) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.005892 (rank : 123) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.019112 (rank : 93) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.037265 (rank : 69) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.004718 (rank : 125) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.019309 (rank : 92) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
DAXX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.006847 (rank : 120) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.031289 (rank : 74) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
M3K1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.000365 (rank : 131) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53349, Q60831, Q9R0U3, Q9R256 | Gene names | Map3k1, Mekk, Mekk1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 1 (EC 2.7.11.25) (MAPK/ERK kinase kinase 1) (MEK kinase 1) (MEKK 1). | |||||
|
MPPD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.006914 (rank : 118) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15442, Q8N361 | Gene names | MPPED1, C22orf1, FAM1A | |||
|
Domain Architecture |
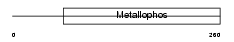
|
|||||
| Description | Metallophosphoesterase domain-containing protein 1 (Adult brain protein 239) (239AB). | |||||
|
MPPD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.006909 (rank : 119) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZG2, Q3TYC7 | Gene names | Mpped1, D15Bwg0669e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallophosphoesterase domain-containing protein 1. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.025069 (rank : 83) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.011789 (rank : 108) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.024720 (rank : 84) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.008593 (rank : 115) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
CT152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.081805 (rank : 40) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.063490 (rank : 47) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.063039 (rank : 48) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.061148 (rank : 50) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.059268 (rank : 51) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053767 (rank : 55) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.062477 (rank : 49) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058591 (rank : 52) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.065089 (rank : 46) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052653 (rank : 56) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.097842 (rank : 39) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052349 (rank : 57) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.050570 (rank : 59) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054778 (rank : 54) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.057258 (rank : 53) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.990656 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.988545 (rank : 3) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.988166 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.974583 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.973135 (rank : 6) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.970029 (rank : 7) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.964151 (rank : 8) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.868271 (rank : 9) | θ value | 1.28137e-46 (rank : 9) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.861301 (rank : 10) | θ value | 8.59492e-43 (rank : 12) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.857454 (rank : 11) | θ value | 1.32601e-43 (rank : 11) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.856005 (rank : 12) | θ value | 2.04573e-44 (rank : 10) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.854856 (rank : 13) | θ value | 1.79215e-40 (rank : 13) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.836323 (rank : 14) | θ value | 1.16434e-31 (rank : 26) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.829583 (rank : 15) | θ value | 3.27365e-34 (rank : 23) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.820264 (rank : 16) | θ value | 1.85889e-29 (rank : 27) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.812057 (rank : 17) | θ value | 1.98146e-39 (rank : 14) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.808720 (rank : 18) | θ value | 2.86122e-38 (rank : 16) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.808160 (rank : 19) | θ value | 2.19075e-38 (rank : 15) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.807813 (rank : 20) | θ value | 2.86122e-38 (rank : 17) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.802843 (rank : 21) | θ value | 5.96599e-36 (rank : 20) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.799174 (rank : 22) | θ value | 4.88048e-38 (rank : 19) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.798895 (rank : 23) | θ value | 4.88048e-38 (rank : 18) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.798329 (rank : 24) | θ value | 3.61944e-33 (rank : 24) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.793221 (rank : 25) | θ value | 6.82597e-32 (rank : 25) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.792858 (rank : 26) | θ value | 8.61488e-35 (rank : 22) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.792658 (rank : 27) | θ value | 8.61488e-35 (rank : 21) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.781330 (rank : 28) | θ value | 2.42779e-29 (rank : 28) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.781208 (rank : 29) | θ value | 2.42779e-29 (rank : 29) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.301563 (rank : 30) | θ value | 1.87187e-05 (rank : 30) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.271079 (rank : 31) | θ value | 0.0193708 (rank : 45) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.264236 (rank : 32) | θ value | 0.00869519 (rank : 42) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.261150 (rank : 33) | θ value | 0.00869519 (rank : 43) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.246775 (rank : 34) | θ value | 0.00390308 (rank : 39) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.242676 (rank : 35) | θ value | 0.00390308 (rank : 38) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.238765 (rank : 36) | θ value | 0.00175202 (rank : 34) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.233340 (rank : 37) | θ value | 0.00175202 (rank : 33) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.147806 (rank : 38) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.097842 (rank : 39) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
CT152_HUMAN
|
||||||
| NC score | 0.081805 (rank : 40) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.073591 (rank : 41) | θ value | 0.00390308 (rank : 40) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.073587 (rank : 42) | θ value | 0.00228821 (rank : 37) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.073335 (rank : 43) | θ value | 0.00228821 (rank : 36) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP2_MOUSE
|
||||||
| NC score | 0.072640 (rank : 44) | θ value | 4.1701e-05 (rank : 31) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.072576 (rank : 45) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.065089 (rank : 46) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.063490 (rank : 47) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.063039 (rank : 48) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.062477 (rank : 49) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.061148 (rank : 50) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.059268 (rank : 51) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.058591 (rank : 52) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.057258 (rank : 53) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.054778 (rank : 54) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.053767 (rank : 55) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.052653 (rank : 56) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.052349 (rank : 57) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.051719 (rank : 58) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.050570 (rank : 59) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.050173 (rank : 60) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.047089 (rank : 61) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.043498 (rank : 62) | θ value | 0.00665767 (rank : 41) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.043069 (rank : 63) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
DCP1B_HUMAN
|
||||||
| NC score | 0.042134 (rank : 64) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.041390 (rank : 65) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
PP14A_MOUSE
|
||||||
| NC score | 0.039423 (rank : 66) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VC7, Q99MB9 | Gene names | Ppp1r14a, Cpi17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14A (17 kDa PKC-potentiated inhibitory protein of PP1) (CPI-17). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.038492 (rank : 67) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.038194 (rank : 68) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.037265 (rank : 69) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.035371 (rank : 70) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.035332 (rank : 71) | θ value | 0.00175202 (rank : 35) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.034941 (rank : 72) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.031375 (rank : 73) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.031289 (rank : 74) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.030254 (rank : 75) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.029397 (rank : 76) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.028517 (rank : 77) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.028283 (rank : 78) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.027998 (rank : 79) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.027030 (rank : 80) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.025395 (rank : 81) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.025272 (rank : 82) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.025069 (rank : 83) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.024720 (rank : 84) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.023079 (rank : 85) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.022766 (rank : 86) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.022275 (rank : 87) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.022060 (rank : 88) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.021725 (rank : 89) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
PRGC1_MOUSE
|
||||||
| NC score | 0.021331 (rank : 90) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.020931 (rank : 91) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.019309 (rank : 92) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.019112 (rank : 93) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.018458 (rank : 94) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.017732 (rank : 95) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.017498 (rank : 96) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.017097 (rank : 97) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.016928 (rank : 98) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
CCR3_MOUSE
|
||||||
| NC score | 0.016526 (rank : 99) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51678 | Gene names | Ccr3, Cmkbr1l2, Cmkbr3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable C-C chemokine receptor type 3 (C-C CKR-3) (CC-CKR-3) (CCR-3) (CCR3) (CKR3) (Macrophage inflammatory protein 1-alpha receptor-like 2) (MIP-1 alpha RL2) (CD193 antigen). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.016491 (rank : 100) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.015636 (rank : 101) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.014898 (rank : 102) | θ value | 0.0736092 (rank : 46) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.014564 (rank : 103) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.014379 (rank : 104) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.013469 (rank : 105) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RL4_HUMAN
|
||||||
| NC score | 0.012912 (rank : 106) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36578, P39029, Q4VBR0, Q969Z9 | Gene names | RPL4, RPL1 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L4 (L1). | |||||
|
SCYL2_MOUSE
|
||||||
| NC score | 0.012864 (rank : 107) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.011789 (rank : 108) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
PRPF3_MOUSE
|
||||||
| NC score | 0.011373 (rank : 109) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
HRSL5_MOUSE
|
||||||
| NC score | 0.010568 (rank : 110) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPX5, Q9CVQ2, Q9CVS0 | Gene names | Hrasls5, Hrlp5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
CAH11_HUMAN
|
||||||
| NC score | 0.009948 (rank : 111) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75493, O60596, Q9UEC4 | Gene names | CA11, CARP2 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase-related protein 2 precursor (CARP-2) (CA-RP II) (CA-XI) (Carbonic anhydrase-related protein 11) (CARP XI) (CA-RP XI). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.009452 (rank : 112) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.009413 (rank : 113) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
CAH11_MOUSE
|
||||||
| NC score | 0.008729 (rank : 114) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70354, Q8VEA9 | Gene names | Ca11, Car11, Carp2 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase-related protein 2 precursor (CARP-2) (CA-RP II) (CA-XI) (Carbonic anhydrase-related protein 11) (CARP XI) (CA-RP XI). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.008593 (rank : 115) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
GCP6_HUMAN
|
||||||
| NC score | 0.008442 (rank : 116) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.008407 (rank : 117) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
MPPD1_HUMAN
|
||||||
| NC score | 0.006914 (rank : 118) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15442, Q8N361 | Gene names | MPPED1, C22orf1, FAM1A | |||
|
Domain Architecture |

|
|||||
| Description | Metallophosphoesterase domain-containing protein 1 (Adult brain protein 239) (239AB). | |||||
|
MPPD1_MOUSE
|
||||||
| NC score | 0.006909 (rank : 119) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZG2, Q3TYC7 | Gene names | Mpped1, D15Bwg0669e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallophosphoesterase domain-containing protein 1. | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.006847 (rank : 120) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
ELL_MOUSE
|
||||||
| NC score | 0.006697 (rank : 121) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
SIG10_HUMAN
|
||||||
| NC score | 0.006173 (rank : 122) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.005892 (rank : 123) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
MYPN_HUMAN
|
||||||
| NC score | 0.004871 (rank : 124) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.004718 (rank : 125) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.003779 (rank : 126) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.003543 (rank : 127) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.002805 (rank : 128) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
TAAR1_HUMAN
|
||||||
| NC score | 0.002737 (rank : 129) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RJ0, Q3MIH8, Q5VUQ1 | Gene names | TAAR1, TA1, TAR1, TRAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Trace amine-associated receptor 1 (Trace amine receptor 1) (TaR-1). | |||||
|
PC11X_HUMAN
|
||||||
| NC score | 0.002206 (rank : 130) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
M3K1_MOUSE
|
||||||
| NC score | 0.000365 (rank : 131) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53349, Q60831, Q9R0U3, Q9R256 | Gene names | Map3k1, Mekk, Mekk1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 1 (EC 2.7.11.25) (MAPK/ERK kinase kinase 1) (MEK kinase 1) (MEKK 1). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.000150 (rank : 132) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
HMX3_MOUSE
|
||||||
| NC score | -0.000792 (rank : 133) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42581 | Gene names | Hmx3, Nkx-5.1, Nkx5-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HMX3 (Homeobox protein Nkx-5.1). | |||||
|
ZN707_HUMAN
|
||||||
| NC score | -0.004003 (rank : 134) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96C28 | Gene names | ZNF707 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 707. | |||||
|
ZN500_HUMAN
|
||||||
| NC score | -0.004326 (rank : 135) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 500. | |||||