Please be patient as the page loads
|
HCN1_MOUSE
|
||||||
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HCN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990656 (rank : 2) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 172 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.960621 (rank : 8) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.970705 (rank : 5) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.984889 (rank : 4) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.985210 (rank : 3) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.969226 (rank : 6) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.967669 (rank : 7) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 9) | NC score | 0.864423 (rank : 9) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 2.67181e-44 (rank : 10) | NC score | 0.852750 (rank : 12) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 11) | NC score | 0.853621 (rank : 11) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | 8.59492e-43 (rank : 12) | NC score | 0.857414 (rank : 10) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 13) | NC score | 0.851416 (rank : 13) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 14) | NC score | 0.808345 (rank : 17) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 15) | NC score | 0.804661 (rank : 19) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 2.19075e-38 (rank : 16) | NC score | 0.795328 (rank : 23) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 17) | NC score | 0.795665 (rank : 22) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 18) | NC score | 0.804862 (rank : 18) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 19) | NC score | 0.804142 (rank : 20) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 20) | NC score | 0.799298 (rank : 21) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 21) | NC score | 0.789071 (rank : 27) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 8.61488e-35 (rank : 22) | NC score | 0.789243 (rank : 26) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 23) | NC score | 0.825958 (rank : 15) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 24) | NC score | 0.794930 (rank : 24) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 25) | NC score | 0.789504 (rank : 25) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 26) | NC score | 0.832592 (rank : 14) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 27) | NC score | 0.816657 (rank : 16) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 28) | NC score | 0.777668 (rank : 28) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 29) | NC score | 0.777258 (rank : 29) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 30) | NC score | 0.299784 (rank : 30) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KGP2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 31) | NC score | 0.075050 (rank : 44) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.074911 (rank : 45) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KAP3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 33) | NC score | 0.231562 (rank : 37) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 34) | NC score | 0.236955 (rank : 36) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 35) | NC score | 0.075329 (rank : 43) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 36) | NC score | 0.075569 (rank : 42) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KAP2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 37) | NC score | 0.241766 (rank : 35) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 38) | NC score | 0.244998 (rank : 34) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 39) | NC score | 0.075570 (rank : 41) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KAP1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 40) | NC score | 0.262424 (rank : 32) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 41) | NC score | 0.259359 (rank : 33) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 42) | NC score | 0.057254 (rank : 53) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
CCR3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.016075 (rank : 108) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51678 | Gene names | Ccr3, Cmkbr1l2, Cmkbr3 | |||
|
Domain Architecture |
|
|||||
| Description | Probable C-C chemokine receptor type 3 (C-C CKR-3) (CC-CKR-3) (CCR-3) (CCR3) (CKR3) (Macrophage inflammatory protein 1-alpha receptor-like 2) (MIP-1 alpha RL2) (CD193 antigen). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 44) | NC score | 0.269155 (rank : 31) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
INVO_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.035666 (rank : 75) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 46) | NC score | 0.028783 (rank : 82) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.040273 (rank : 66) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.023298 (rank : 93) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.020584 (rank : 98) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 50) | NC score | 0.012236 (rank : 133) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 51) | NC score | 0.012544 (rank : 132) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
UBP2L_HUMAN
|
||||||
| θ value | 0.125558 (rank : 52) | NC score | 0.039190 (rank : 68) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 53) | NC score | 0.028034 (rank : 85) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.040789 (rank : 64) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 0.21417 (rank : 55) | NC score | 0.042889 (rank : 63) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
SYPH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.020445 (rank : 99) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08247, Q6P2F7 | Gene names | SYP | |||
|
Domain Architecture |
|
|||||
| Description | Synaptophysin (Major synaptic vesicle protein p38). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 57) | NC score | 0.016998 (rank : 107) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 58) | NC score | 0.012797 (rank : 129) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 59) | NC score | 0.033318 (rank : 78) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | 0.025062 (rank : 89) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
RING1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 61) | NC score | 0.018022 (rank : 102) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.365318 (rank : 62) | NC score | 0.020798 (rank : 97) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
DCP1B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.039655 (rank : 67) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.022646 (rank : 95) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 65) | NC score | 0.023320 (rank : 92) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 66) | NC score | 0.023679 (rank : 91) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 67) | NC score | 0.035743 (rank : 74) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 68) | NC score | 0.020935 (rank : 96) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP2L_MOUSE
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.035412 (rank : 76) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
AMOL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.017730 (rank : 104) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 71) | NC score | 0.031915 (rank : 79) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
EVX2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 72) | NC score | 0.007477 (rank : 161) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03828 | Gene names | EVX2 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox even-skipped homolog protein 2 (EVX-2). | |||||
|
EVX2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 73) | NC score | 0.007511 (rank : 160) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49749 | Gene names | Evx2, Evx-2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 2 (EVX-2). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.014184 (rank : 121) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.053887 (rank : 56) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 0.813845 (rank : 76) | NC score | 0.040334 (rank : 65) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.011210 (rank : 142) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.014261 (rank : 120) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.026590 (rank : 87) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
HAP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.024602 (rank : 90) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |
|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.020338 (rank : 100) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.012196 (rank : 135) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.017271 (rank : 105) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.013602 (rank : 125) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CT152_MOUSE
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.146894 (rank : 38) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.003326 (rank : 177) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.015637 (rank : 111) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.037576 (rank : 72) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 89) | NC score | 0.038291 (rank : 70) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.011580 (rank : 140) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.001801 (rank : 182) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.012226 (rank : 134) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.010790 (rank : 144) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.033498 (rank : 77) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.012048 (rank : 137) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
LARP4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.013828 (rank : 123) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71RC2, Q5CZ97, Q96NF9 | Gene names | LARP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
NKX32_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.003652 (rank : 176) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78367 | Gene names | BAPX1, NKX3B | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein Nkx-3.2 (Bagpipe homeobox protein homolog 1). | |||||
|
OSB10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.008059 (rank : 157) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.015361 (rank : 113) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.007214 (rank : 162) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.053383 (rank : 57) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.006974 (rank : 164) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.006437 (rank : 167) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
LAP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.001886 (rank : 180) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.006047 (rank : 170) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.012915 (rank : 128) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.036898 (rank : 73) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.017897 (rank : 103) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
GAK18_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.004319 (rank : 173) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
LORI_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.025862 (rank : 88) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.014659 (rank : 117) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.010491 (rank : 149) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.052349 (rank : 60) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.014999 (rank : 115) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
PP14A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.038377 (rank : 69) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VC7, Q99MB9 | Gene names | Ppp1r14a, Cpi17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14A (17 kDa PKC-potentiated inhibitory protein of PP1) (CPI-17). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.012919 (rank : 127) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TAAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.002202 (rank : 179) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RJ0, Q3MIH8, Q5VUQ1 | Gene names | TAAR1, TA1, TAR1, TRAR1 | |||
|
Domain Architecture |
|
|||||
| Description | Trace amine-associated receptor 1 (Trace amine receptor 1) (TaR-1). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.008449 (rank : 156) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.015329 (rank : 114) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
VCIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.015838 (rank : 110) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
AMOL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.015849 (rank : 109) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.015392 (rank : 112) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.012176 (rank : 136) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.004764 (rank : 171) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
E2F3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.006175 (rank : 169) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |
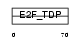
|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
FOXD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.001146 (rank : 184) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |
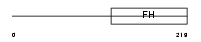
|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.012775 (rank : 130) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.018388 (rank : 101) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.010014 (rank : 153) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SYPH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.014298 (rank : 119) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62277, Q8BRQ0, Q91WI8 | Gene names | Syp | |||
|
Domain Architecture |
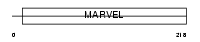
|
|||||
| Description | Synaptophysin (Major synaptic vesicle protein p38) (BM89 antigen). | |||||
|
ANGL4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.002901 (rank : 178) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1P8, Q78ZJ9, Q9JHX7, Q9JLX7 | Gene names | Angptl4, Farp, Fiaf, Ng27 | |||
|
Domain Architecture |
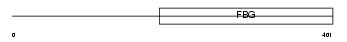
|
|||||
| Description | Angiopoietin-related protein 4 precursor (Angiopoietin-like 4) (Hepatic fibrinogen/angiopoietin-related protein) (HFARP) (Fasting- induced adipose factor) (Secreted protein Bk89) (425O18-1). | |||||
|
AR13B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.006382 (rank : 168) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
BL1S3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.013546 (rank : 126) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5U5M8, Q8C6R4, Q8R0Z3 | Gene names | Bloc1s3, Blos3, Rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Biogenesis of lysosome-related organelles complex-1 subunit 3 (Reduced pigmentation protein) (BLOC subunit 3). | |||||
|
CD45_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.001808 (rank : 181) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
DLX5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.001320 (rank : 183) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.007799 (rank : 159) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.029307 (rank : 81) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MAML1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.028216 (rank : 83) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.010098 (rank : 152) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.011668 (rank : 139) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.010514 (rank : 148) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.010535 (rank : 147) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.013925 (rank : 122) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
VCIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.014556 (rank : 118) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.023284 (rank : 94) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.010274 (rank : 151) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
FES_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.001019 (rank : 185) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.027868 (rank : 86) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.052931 (rank : 59) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
OXR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.014784 (rank : 116) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.010761 (rank : 145) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.037641 (rank : 71) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.006553 (rank : 166) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ATRIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.007038 (rank : 163) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
BCL6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | -0.005538 (rank : 186) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.013724 (rank : 124) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.028087 (rank : 84) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.010610 (rank : 146) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.003692 (rank : 175) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
GCC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.011225 (rank : 141) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.010426 (rank : 150) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.011172 (rank : 143) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.031067 (rank : 80) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.012744 (rank : 131) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KIFC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.009354 (rank : 154) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
KIFC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.009002 (rank : 155) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 571 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35231, Q91YQ2 | Gene names | Kifc3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.006608 (rank : 165) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.008049 (rank : 158) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
PRGC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.017110 (rank : 106) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.011841 (rank : 138) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.004575 (rank : 172) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
UBIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.003758 (rank : 174) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
CT152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.081147 (rank : 40) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.063019 (rank : 47) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.062568 (rank : 48) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.060683 (rank : 50) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.058813 (rank : 51) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053270 (rank : 58) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.062022 (rank : 49) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.058288 (rank : 52) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.064585 (rank : 46) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052248 (rank : 61) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.097153 (rank : 39) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.050089 (rank : 62) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.054278 (rank : 55) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.056694 (rank : 54) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 172 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.990656 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.985210 (rank : 3) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.984889 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.970705 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.969226 (rank : 6) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.967669 (rank : 7) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.960621 (rank : 8) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.864423 (rank : 9) | θ value | 1.28137e-46 (rank : 9) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.857414 (rank : 10) | θ value | 8.59492e-43 (rank : 12) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.853621 (rank : 11) | θ value | 1.32601e-43 (rank : 11) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.852750 (rank : 12) | θ value | 2.67181e-44 (rank : 10) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.851416 (rank : 13) | θ value | 1.79215e-40 (rank : 13) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.832592 (rank : 14) | θ value | 1.16434e-31 (rank : 26) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.825958 (rank : 15) | θ value | 2.50655e-34 (rank : 23) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.816657 (rank : 16) | θ value | 1.85889e-29 (rank : 27) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.808345 (rank : 17) | θ value | 1.98146e-39 (rank : 14) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.804862 (rank : 18) | θ value | 2.86122e-38 (rank : 18) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.804661 (rank : 19) | θ value | 2.19075e-38 (rank : 15) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.804142 (rank : 20) | θ value | 2.86122e-38 (rank : 19) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.799298 (rank : 21) | θ value | 5.96599e-36 (rank : 20) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.795665 (rank : 22) | θ value | 2.19075e-38 (rank : 17) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.795328 (rank : 23) | θ value | 2.19075e-38 (rank : 16) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.794930 (rank : 24) | θ value | 3.61944e-33 (rank : 24) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.789504 (rank : 25) | θ value | 5.22648e-32 (rank : 25) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.789243 (rank : 26) | θ value | 8.61488e-35 (rank : 22) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.789071 (rank : 27) | θ value | 8.61488e-35 (rank : 21) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.777668 (rank : 28) | θ value | 2.42779e-29 (rank : 28) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.777258 (rank : 29) | θ value | 2.42779e-29 (rank : 29) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.299784 (rank : 30) | θ value | 1.87187e-05 (rank : 30) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.269155 (rank : 31) | θ value | 0.0193708 (rank : 44) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.262424 (rank : 32) | θ value | 0.00869519 (rank : 40) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.259359 (rank : 33) | θ value | 0.00869519 (rank : 41) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.244998 (rank : 34) | θ value | 0.00390308 (rank : 38) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.241766 (rank : 35) | θ value | 0.00390308 (rank : 37) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.236955 (rank : 36) | θ value | 0.00228821 (rank : 34) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.231562 (rank : 37) | θ value | 0.00228821 (rank : 33) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.146894 (rank : 38) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.097153 (rank : 39) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
CT152_HUMAN
|
||||||
| NC score | 0.081147 (rank : 40) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M20, Q5T3S1, Q9BR36, Q9BWY5 | Gene names | C20orf152 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152. | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.075570 (rank : 41) | θ value | 0.00390308 (rank : 39) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.075569 (rank : 42) | θ value | 0.00228821 (rank : 36) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.075329 (rank : 43) | θ value | 0.00228821 (rank : 35) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP2_MOUSE
|
||||||
| NC score | 0.075050 (rank : 44) | θ value | 5.44631e-05 (rank : 31) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.074911 (rank : 45) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.064585 (rank : 46) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.063019 (rank : 47) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.062568 (rank : 48) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.062022 (rank : 49) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.060683 (rank : 50) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.058813 (rank : 51) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.058288 (rank : 52) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.057254 (rank : 53) | θ value | 0.00869519 (rank : 42) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.056694 (rank : 54) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.054278 (rank : 55) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.053887 (rank : 56) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.053383 (rank : 57) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.053270 (rank : 58) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.052931 (rank : 59) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.052349 (rank : 60) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.052248 (rank : 61) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.050089 (rank : 62) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.042889 (rank : 63) | θ value | 0.21417 (rank : 55) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.040789 (rank : 64) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.040334 (rank : 65) | θ value | 0.813845 (rank : 76) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.040273 (rank : 66) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
DCP1B_HUMAN
|
||||||
| NC score | 0.039655 (rank : 67) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
UBP2L_HUMAN
|
||||||
| NC score | 0.039190 (rank : 68) | θ value | 0.125558 (rank : 52) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
PP14A_MOUSE
|
||||||
| NC score | 0.038377 (rank : 69) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VC7, Q99MB9 | Gene names | Ppp1r14a, Cpi17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14A (17 kDa PKC-potentiated inhibitory protein of PP1) (CPI-17). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.038291 (rank : 70) | θ value | 1.38821 (rank : 89) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.037641 (rank : 71) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.037576 (rank : 72) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.036898 (rank : 73) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.035743 (rank : 74) | θ value | 0.47712 (rank : 67) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.035666 (rank : 75) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
UBP2L_MOUSE
|
||||||
| NC score | 0.035412 (rank : 76) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.033498 (rank : 77) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.033318 (rank : 78) | θ value | 0.279714 (rank : 59) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.031915 (rank : 79) | θ value | 0.62314 (rank : 71) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.031067 (rank : 80) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.029307 (rank : 81) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.028783 (rank : 82) | θ value | 0.0252991 (rank : 46) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.028216 (rank : 83) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.028087 (rank : 84) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.028034 (rank : 85) | θ value | 0.163984 (rank : 53) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.027868 (rank : 86) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.026590 (rank : 87) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.025862 (rank : 88) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.025062 (rank : 89) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
HAP1_MOUSE
|
||||||
| NC score | 0.024602 (rank : 90) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.023679 (rank : 91) | θ value | 0.47712 (rank : 66) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.023320 (rank : 92) | θ value | 0.47712 (rank : 65) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.023298 (rank : 93) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.023284 (rank : 94) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.022646 (rank : 95) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.020935 (rank : 96) | θ value | 0.47712 (rank : 68) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.020798 (rank : 97) | θ value | 0.365318 (rank : 62) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.020584 (rank : 98) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
SYPH_HUMAN
|
||||||
| NC score | 0.020445 (rank : 99) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08247, Q6P2F7 | Gene names | SYP | |||
|
Domain Architecture |

|
|||||
| Description | Synaptophysin (Major synaptic vesicle protein p38). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.020338 (rank : 100) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.018388 (rank : 101) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
RING1_HUMAN
|
||||||
| NC score | 0.018022 (rank : 102) | θ value | 0.365318 (rank : 61) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.017897 (rank : 103) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
AMOL1_MOUSE
|
||||||
| NC score | 0.017730 (rank : 104) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.017271 (rank : 105) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
PRGC1_MOUSE
|
||||||
| NC score | 0.017110 (rank : 106) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.016998 (rank : 107) | θ value | 0.279714 (rank : 57) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
CCR3_MOUSE
|
||||||
| NC score | 0.016075 (rank : 108) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51678 | Gene names | Ccr3, Cmkbr1l2, Cmkbr3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable C-C chemokine receptor type 3 (C-C CKR-3) (CC-CKR-3) (CCR-3) (CCR3) (CKR3) (Macrophage inflammatory protein 1-alpha receptor-like 2) (MIP-1 alpha RL2) (CD193 antigen). | |||||
|
AMOL1_HUMAN
|
||||||
| NC score | 0.015849 (rank : 109) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
VCIP1_HUMAN
|
||||||
| NC score | 0.015838 (rank : 110) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.015637 (rank : 111) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.015392 (rank : 112) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.015361 (rank : 113) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.015329 (rank : 114) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.014999 (rank : 115) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
OXR1_HUMAN
|
||||||
| NC score | 0.014784 (rank : 116) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.014659 (rank : 117) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
VCIP1_MOUSE
|
||||||
| NC score | 0.014556 (rank : 118) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
SYPH_MOUSE
|
||||||
| NC score | 0.014298 (rank : 119) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62277, Q8BRQ0, Q91WI8 | Gene names | Syp | |||
|
Domain Architecture |
|
|||||
| Description | Synaptophysin (Major synaptic vesicle protein p38) (BM89 antigen). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.014261 (rank : 120) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.014184 (rank : 121) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.013925 (rank : 122) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
LARP4_HUMAN
|
||||||
| NC score | 0.013828 (rank : 123) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71RC2, Q5CZ97, Q96NF9 | Gene names | LARP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.013724 (rank : 124) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.013602 (rank : 125) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
BL1S3_MOUSE
|
||||||
| NC score | 0.013546 (rank : 126) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5U5M8, Q8C6R4, Q8R0Z3 | Gene names | Bloc1s3, Blos3, Rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Biogenesis of lysosome-related organelles complex-1 subunit 3 (Reduced pigmentation protein) (BLOC subunit 3). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.012919 (rank : 127) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.012915 (rank : 128) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.012797 (rank : 129) | θ value | 0.279714 (rank : 58) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.012775 (rank : 130) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.012744 (rank : 131) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.012544 (rank : 132) | θ value | 0.0961366 (rank : 51) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.012236 (rank : 133) | θ value | 0.0961366 (rank : 50) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.012226 (rank : 134) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.012196 (rank : 135) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.012176 (rank : 136) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.012048 (rank : 137) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.011841 (rank : 138) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.011668 (rank : 139) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.011580 (rank : 140) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
GCC1_HUMAN
|
||||||
| NC score | 0.011225 (rank : 141) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.011210 (rank : 142) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.011172 (rank : 143) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.010790 (rank : 144) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.010761 (rank : 145) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.010610 (rank : 146) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.010535 (rank : 147) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.010514 (rank : 148) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.010491 (rank : 149) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.010426 (rank : 150) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.010274 (rank : 151) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.010098 (rank : 152) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.010014 (rank : 153) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
KIFC3_HUMAN
|
||||||
| NC score | 0.009354 (rank : 154) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
KIFC3_MOUSE
|
||||||
| NC score | 0.009002 (rank : 155) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 571 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35231, Q91YQ2 | Gene names | Kifc3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.008449 (rank : 156) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
OSB10_HUMAN
|
||||||
| NC score | 0.008059 (rank : 157) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.008049 (rank : 158) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.007799 (rank : 159) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
EVX2_MOUSE
|
||||||
| NC score | 0.007511 (rank : 160) | θ value | 0.62314 (rank : 73) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49749 | Gene names | Evx2, Evx-2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 2 (EVX-2). | |||||
|
EVX2_HUMAN
|
||||||
| NC score | 0.007477 (rank : 161) | θ value | 0.62314 (rank : 72) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03828 | Gene names | EVX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 2 (EVX-2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.007214 (rank : 162) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ATRIP_MOUSE
|
||||||
| NC score | 0.007038 (rank : 163) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.006974 (rank : 164) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.006608 (rank : 165) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.006553 (rank : 166) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.006437 (rank : 167) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
AR13B_HUMAN
|
||||||
| NC score | 0.006382 (rank : 168) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
E2F3_HUMAN
|
||||||
| NC score | 0.006175 (rank : 169) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.006047 (rank : 170) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.004764 (rank : 171) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.004575 (rank : 172) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.004319 (rank : 173) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
UBIP1_MOUSE
|
||||||
| NC score | 0.003758 (rank : 174) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.003692 (rank : 175) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
NKX32_HUMAN
|
||||||
| NC score | 0.003652 (rank : 176) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78367 | Gene names | BAPX1, NKX3B | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.2 (Bagpipe homeobox protein homolog 1). | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.003326 (rank : 177) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
ANGL4_MOUSE
|
||||||
| NC score | 0.002901 (rank : 178) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1P8, Q78ZJ9, Q9JHX7, Q9JLX7 | Gene names | Angptl4, Farp, Fiaf, Ng27 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 4 precursor (Angiopoietin-like 4) (Hepatic fibrinogen/angiopoietin-related protein) (HFARP) (Fasting- induced adipose factor) (Secreted protein Bk89) (425O18-1). | |||||
|
TAAR1_HUMAN
|
||||||
| NC score | 0.002202 (rank : 179) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RJ0, Q3MIH8, Q5VUQ1 | Gene names | TAAR1, TA1, TAR1, TRAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Trace amine-associated receptor 1 (Trace amine receptor 1) (TaR-1). | |||||
|
LAP2_HUMAN
|
||||||
| NC score | 0.001886 (rank : 180) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.001808 (rank : 181) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.001801 (rank : 182) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
DLX5_HUMAN
|
||||||
| NC score | 0.001320 (rank : 183) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
FOXD2_HUMAN
|
||||||
| NC score | 0.001146 (rank : 184) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
FES_HUMAN
|
||||||
| NC score | 0.001019 (rank : 185) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
BCL6_MOUSE
|
||||||
| NC score | -0.005538 (rank : 186) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 172 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||