Please be patient as the page loads
|
HTRA4_HUMAN
|
||||||
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HTRA4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | 6.85399e-133 (rank : 2) | NC score | 0.946024 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 1.29261e-131 (rank : 3) | NC score | 0.947567 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | 1.09934e-114 (rank : 4) | NC score | 0.957509 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
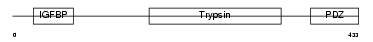
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 9.30652e-114 (rank : 5) | NC score | 0.956735 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
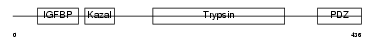
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA2_MOUSE
|
||||||
| θ value | 6.73165e-80 (rank : 6) | NC score | 0.911642 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |
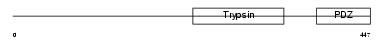
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_HUMAN
|
||||||
| θ value | 1.49966e-79 (rank : 7) | NC score | 0.910634 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |
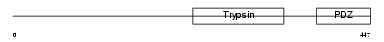
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
IBP7_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 8) | NC score | 0.442611 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
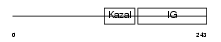
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 9) | NC score | 0.439981 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
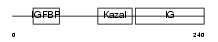
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
ESM1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 10) | NC score | 0.355661 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
NOV_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 11) | NC score | 0.273916 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |
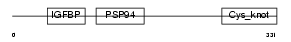
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
NOV_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 12) | NC score | 0.265940 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |
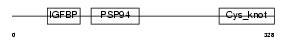
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
WISP2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 13) | NC score | 0.281926 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
ESM1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 14) | NC score | 0.374749 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |
|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
CTGF_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 15) | NC score | 0.273370 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |
|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
CYR61_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 16) | NC score | 0.276336 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
CYR61_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 17) | NC score | 0.276882 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
CTGF_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 18) | NC score | 0.243779 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |
|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 19) | NC score | 0.336980 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
WISP2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.246864 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |
|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 21) | NC score | 0.352713 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.099793 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.075723 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.065863 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.254659 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
PDZD7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.081698 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.074963 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.127482 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
SLIT1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.016390 (rank : 134) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
WISP3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.248787 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.283922 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.308268 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.125308 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.082182 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
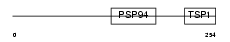
WISP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.204720 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |
|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
WISP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.214495 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
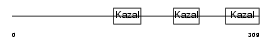
FST_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.108575 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.065036 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
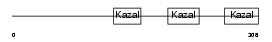
FST_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.107763 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.176116 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.176617 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
ISK5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.093647 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
KRA24_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.035290 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
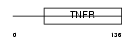
TNR14_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.035262 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.058397 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.175968 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.156675 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.138330 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
RHPN1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.083036 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
IPK1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.135539 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |
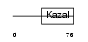
|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.156080 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.037314 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.037408 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.176540 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.175070 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
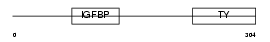
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
ISK3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.154031 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |
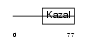
|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.136097 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.143125 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.043355 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
NOS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.042706 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
RHPN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.081569 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.085575 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.086617 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
IBP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.194866 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
PDZD3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.098739 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
TX1B3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.088432 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.087915 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
GRASP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.069859 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
GRASP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.066096 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.051508 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.032995 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.033214 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.013316 (rank : 137) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.015263 (rank : 136) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
CHRD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.017761 (rank : 133) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0E2 | Gene names | Chrd | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.162289 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.116463 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
KR107_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.028091 (rank : 126) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.105451 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.048137 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.052928 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.043343 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
CFAI_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.002383 (rank : 143) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.031298 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.031597 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
KR108_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.037942 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.244173 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.037908 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
PSCBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.059617 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
SEM4G_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.003778 (rank : 139) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
ZO2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.040613 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.039476 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.042424 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
EMR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.005034 (rank : 138) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.003708 (rank : 141) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.003725 (rank : 140) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NOS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.031613 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z0J4, Q64208 | Gene names | Nos1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
PDZD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.102128 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.075866 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.071683 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ZO3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.062026 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.023502 (rank : 131) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.023507 (rank : 130) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.030342 (rank : 125) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.121197 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.031289 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.031067 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.148020 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
KR106_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.031916 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR171_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.035340 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.045265 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.098446 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.003300 (rank : 142) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
NFX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.024687 (rank : 129) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
PAR6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.026488 (rank : 128) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYG5, Q9Y510 | Gene names | PARD6B, PAR6B | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
PAR6B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.027179 (rank : 127) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JK83, Q8R3J8 | Gene names | Pard6b, Par6b | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.018766 (rank : 132) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
RECK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.062530 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.015598 (rank : 135) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
FSTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.056683 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
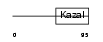
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.055893 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.135226 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.126975 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.122966 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.068173 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.064824 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
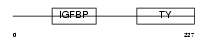
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
ISK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050862 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ISK6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.069666 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.081768 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.051154 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
NHERF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.092119 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
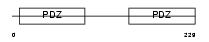
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.088241 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.075502 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.068781 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.123402 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.122009 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.076220 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
PDZK4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.076711 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QY39, Q6PHP2 | Gene names | Pdzk4, Pdzrn4l, Xlu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
RECK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.058725 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
SDCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.054428 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |
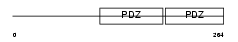
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
SDCB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.050246 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |
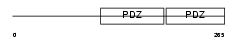
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
TEFF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.065173 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.068169 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.957509 (rank : 2) | θ value | 1.09934e-114 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.956735 (rank : 3) | θ value | 9.30652e-114 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.947567 (rank : 4) | θ value | 1.29261e-131 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.946024 (rank : 5) | θ value | 6.85399e-133 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA2_MOUSE
|
||||||
| NC score | 0.911642 (rank : 6) | θ value | 6.73165e-80 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_HUMAN
|
||||||
| NC score | 0.910634 (rank : 7) | θ value | 1.49966e-79 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.442611 (rank : 8) | θ value | 4.59992e-12 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.439981 (rank : 9) | θ value | 6.64225e-11 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
ESM1_MOUSE
|
||||||
| NC score | 0.374749 (rank : 10) | θ value | 0.000121331 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
ESM1_HUMAN
|
||||||
| NC score | 0.355661 (rank : 11) | θ value | 2.88788e-06 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.352713 (rank : 12) | θ value | 0.00869519 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.336980 (rank : 13) | θ value | 0.00298849 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.308268 (rank : 14) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.283922 (rank : 15) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
WISP2_HUMAN
|
||||||
| NC score | 0.281926 (rank : 16) | θ value | 9.29e-05 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
CYR61_MOUSE
|
||||||
| NC score | 0.276882 (rank : 17) | θ value | 0.00134147 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
CYR61_HUMAN
|
||||||
| NC score | 0.276336 (rank : 18) | θ value | 0.00134147 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
NOV_HUMAN
|
||||||
| NC score | 0.273916 (rank : 19) | θ value | 1.09739e-05 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
CTGF_HUMAN
|
||||||
| NC score | 0.273370 (rank : 20) | θ value | 0.00102713 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
NOV_MOUSE
|
||||||
| NC score | 0.265940 (rank : 21) | θ value | 4.1701e-05 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.254659 (rank : 22) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
WISP3_HUMAN
|
||||||
| NC score | 0.248787 (rank : 23) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
WISP2_MOUSE
|
||||||
| NC score | 0.246864 (rank : 24) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.244173 (rank : 25) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
CTGF_MOUSE
|
||||||
| NC score | 0.243779 (rank : 26) | θ value | 0.00175202 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
WISP1_MOUSE
|
||||||
| NC score | 0.214495 (rank : 27) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
WISP1_HUMAN
|
||||||
| NC score | 0.204720 (rank : 28) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.194866 (rank : 29) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.176617 (rank : 30) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.176540 (rank : 31) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.176116 (rank : 32) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.175968 (rank : 33) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.175070 (rank : 34) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.162289 (rank : 35) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.156675 (rank : 36) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.156080 (rank : 37) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
ISK3_MOUSE
|
||||||
| NC score | 0.154031 (rank : 38) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.148020 (rank : 39) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.143125 (rank : 40) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.138330 (rank : 41) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.136097 (rank : 42) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
IPK1_HUMAN
|
||||||
| NC score | 0.135539 (rank : 43) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.135226 (rank : 44) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.127482 (rank : 45) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.126975 (rank : 46) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.125308 (rank : 47) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.123402 (rank : 48) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.122966 (rank : 49) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.122009 (rank : 50) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.121197 (rank : 51) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.116463 (rank : 52) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.108575 (rank : 53) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.107763 (rank : 54) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.105451 (rank : 55) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
PDZD3_MOUSE
|
||||||
| NC score | 0.102128 (rank : 56) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.099793 (rank : 57) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
PDZD3_HUMAN
|
||||||
| NC score | 0.098739 (rank : 58) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.098446 (rank : 59) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.093647 (rank : 60) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.092119 (rank : 61) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
TX1B3_HUMAN
|
||||||
| NC score | 0.088432 (rank : 62) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.088241 (rank : 63) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
TX1B3_MOUSE
|
||||||
| NC score | 0.087915 (rank : 64) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.086617 (rank : 65) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.085575 (rank : 66) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
RHPN1_MOUSE
|
||||||
| NC score | 0.083036 (rank : 67) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.082182 (rank : 68) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.081768 (rank : 69) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
PDZD7_HUMAN
|
||||||
| NC score | 0.081698 (rank : 70) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
RHPN1_HUMAN
|
||||||
| NC score | 0.081569 (rank : 71) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
PDZK4_MOUSE
|
||||||
| NC score | 0.076711 (rank : 72) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QY39, Q6PHP2 | Gene names | Pdzk4, Pdzrn4l, Xlu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
PDZK4_HUMAN
|
||||||
| NC score | 0.076220 (rank : 73) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.075866 (rank : 74) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.075723 (rank : 75) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.075502 (rank : 76) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.074963 (rank : 77) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.071683 (rank : 78) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
GRASP_HUMAN
|
||||||
| NC score | 0.069859 (rank : 79) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
ISK6_MOUSE
|
||||||
| NC score | 0.069666 (rank : 80) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.068781 (rank : 81) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.068173 (rank : 82) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
TEFF2_MOUSE
|
||||||
| NC score | 0.068169 (rank : 83) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
GRASP_MOUSE
|
||||||
| NC score | 0.066096 (rank : 84) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.065863 (rank : 85) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
TEFF2_HUMAN
|
||||||
| NC score | 0.065173 (rank : 86) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.065036 (rank : 87) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.064824 (rank : 88) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.062530 (rank : 89) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
ZO3_MOUSE
|
||||||
| NC score | 0.062026 (rank : 90) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
PSCBP_HUMAN
|
||||||
| NC score | 0.059617 (rank : 91) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
RECK_MOUSE
|
||||||
| NC score | 0.058725 (rank : 92) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.058397 (rank : 93) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.056683 (rank : 94) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.055893 (rank : 95) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
SDCB1_HUMAN
|
||||||
| NC score | 0.054428 (rank : 96) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.052928 (rank : 97) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.051508 (rank : 98) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.051154 (rank : 99) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
ISK6_HUMAN
|
||||||
| NC score | 0.050862 (rank : 100) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
SDCB1_MOUSE
|
||||||
| NC score | 0.050246 (rank : 101) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.048137 (rank : 102) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.045265 (rank : 103) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.043355 (rank : 104) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.043343 (rank : 105) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
NOS1_HUMAN
|
||||||
| NC score | 0.042706 (rank : 106) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.042424 (rank : 107) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.040613 (rank : 108) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.039476 (rank : 109) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.037942 (rank : 110) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.037908 (rank : 111) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.037408 (rank : 112) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.037314 (rank : 113) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.035340 (rank : 114) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.035290 (rank : 115) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
TNR14_HUMAN
|
||||||
| NC score | 0.035262 (rank : 116) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.033214 (rank : 117) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.032995 (rank : 118) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.031916 (rank : 119) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
NOS1_MOUSE
|
||||||
| NC score | 0.031613 (rank : 120) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z0J4, Q64208 | Gene names | Nos1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.031597 (rank : 121) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.031298 (rank : 122) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.031289 (rank : 123) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.031067 (rank : 124) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.030342 (rank : 125) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.028091 (rank : 126) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
PAR6B_MOUSE
|
||||||
| NC score | 0.027179 (rank : 127) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JK83, Q8R3J8 | Gene names | Pard6b, Par6b | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
PAR6B_HUMAN
|
||||||
| NC score | 0.026488 (rank : 128) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYG5, Q9Y510 | Gene names | PARD6B, PAR6B | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.024687 (rank : 129) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
APBA2_MOUSE
|
||||||
| NC score | 0.023507 (rank : 130) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.023502 (rank : 131) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.018766 (rank : 132) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CHRD_MOUSE
|
||||||
| NC score | 0.017761 (rank : 133) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0E2 | Gene names | Chrd | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
SLIT1_HUMAN
|
||||||
| NC score | 0.016390 (rank : 134) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.015598 (rank : 135) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.015263 (rank : 136) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.013316 (rank : 137) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
EMR1_MOUSE
|
||||||
| NC score | 0.005034 (rank : 138) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
SEM4G_MOUSE
|
||||||
| NC score | 0.003778 (rank : 139) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.003725 (rank : 140) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.003708 (rank : 141) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.003300 (rank : 142) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.002383 (rank : 143) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||