Please be patient as the page loads
|
LORI_HUMAN
|
||||||
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TROP_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 1) | NC score | 0.211723 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
LORI_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 3) | NC score | 0.698669 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
JPH4_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 4) | NC score | 0.356069 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 5) | NC score | 0.353306 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 6) | NC score | 0.215562 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.083012 (rank : 71) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.177554 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
JAG2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.063221 (rank : 114) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
ZN297_MOUSE
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.019192 (rank : 193) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0G7 | Gene names | Znf297, Bing1, Zfp297 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 297 (BING1 protein). | |||||
|
JPH3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.280323 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
CREL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.114513 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.113060 (rank : 37) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
KR131_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.095303 (rank : 59) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IUC0, Q3LI79 | Gene names | KRTAP13-1, KAP13.1, KRTAP13.1 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin-associated protein 13-1 (High sulfur keratin-associated protein 13.1). | |||||
|
JPH3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.280605 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
GP123_HUMAN
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.058370 (rank : 132) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
KRHB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.030362 (rank : 190) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.104581 (rank : 49) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.025862 (rank : 192) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.059768 (rank : 125) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.060795 (rank : 120) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
NU214_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.117943 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.093899 (rank : 61) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.066303 (rank : 104) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
FOXF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.009840 (rank : 194) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12946, Q5FWE5 | Gene names | FOXF1, FKHL5, FREAC1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Forkhead-related activator 1). | |||||
|
KRHB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.026577 (rank : 191) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.047626 (rank : 189) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
STAB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.100604 (rank : 52) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
ALS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.073470 (rank : 86) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.073581 (rank : 84) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ASGR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.050785 (rank : 179) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07306 | Gene names | ASGR1 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin H1). | |||||
|
ASGR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.051914 (rank : 172) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
ASPX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.076480 (rank : 76) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
C209C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.052432 (rank : 169) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
CD209_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.052146 (rank : 171) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
CD302_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.050421 (rank : 183) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CDSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.074752 (rank : 83) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
CDSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.113458 (rank : 35) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CLC10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.050683 (rank : 181) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
CLC4D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.061080 (rank : 119) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
CLC4D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.060031 (rank : 121) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
CLC4E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.051232 (rank : 175) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.054757 (rank : 152) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
CLC4G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.058923 (rank : 130) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
CLC4G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.053607 (rank : 160) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
CLC4K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.056608 (rank : 141) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.051521 (rank : 173) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4M_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.053060 (rank : 164) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.067526 (rank : 102) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.050152 (rank : 187) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.098162 (rank : 54) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.096220 (rank : 58) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.096396 (rank : 57) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.097317 (rank : 56) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.051456 (rank : 174) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.053832 (rank : 159) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.050510 (rank : 182) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.054957 (rank : 149) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.074888 (rank : 82) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
EGFL7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.050891 (rank : 178) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXT5, Q6XD35, Q9DCP5 | Gene names | Egfl7, Megf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 7 precursor (Multiple EGF-like domain protein 7) (Multiple epidermal growth factor-like domain protein 7) (Vascular endothelial statin) (VE-statin) (NOTCH4-like protein) (Zneu1). | |||||
|
ELN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.107074 (rank : 46) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
ELN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.164287 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
FA98A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.082231 (rank : 73) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCA5 | Gene names | FAM98A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
FA98A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.072555 (rank : 89) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TJZ6, Q99KJ2 | Gene names | Fam98a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.059579 (rank : 127) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.059661 (rank : 126) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.057699 (rank : 136) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
FCER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.058313 (rank : 133) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
FCER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.054860 (rank : 150) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
HORN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.137421 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
HORN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.151326 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
HPLN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.112898 (rank : 38) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.112004 (rank : 39) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.113248 (rank : 36) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.114010 (rank : 34) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.116234 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
HPLN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.116408 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
HPLN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.111207 (rank : 40) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
HPLN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.109409 (rank : 41) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |
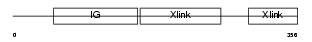
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.055121 (rank : 147) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.053255 (rank : 163) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.057490 (rank : 138) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
JPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.245168 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.244206 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.213292 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.221987 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
K1C9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050740 (rank : 180) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
KR132_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.069257 (rank : 99) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q52LG2 | Gene names | KRTAP13-2, KAP13.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-2. | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.100969 (rank : 51) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR191_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.114737 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUB9, Q3LI75 | Gene names | KRTAP19-1, KAP19.1, KRTAP19.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-1 (High tyrosine-glycine keratin- associated protein 19.1). | |||||
|
KR192_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.093049 (rank : 63) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LHN2 | Gene names | KRTAP19-2, KAP19.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-2. | |||||
|
KR193_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.256075 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
KR195_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.104589 (rank : 48) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI72 | Gene names | KRTAP19-5, KAP19.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-5. | |||||
|
KR197_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.158281 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3SYF9 | Gene names | KRTAP19-7, KAP19.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-7. | |||||
|
KR202_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.275781 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
KR211_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.108264 (rank : 44) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI58 | Gene names | KRTAP21-1, KAP21.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-1. | |||||
|
KR212_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.175201 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3LI59 | Gene names | KRTAP21-2, KAP21.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-2. | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.075512 (rank : 80) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.070670 (rank : 94) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.054775 (rank : 151) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.088190 (rank : 68) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.065211 (rank : 106) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.065948 (rank : 105) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.106905 (rank : 47) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.075831 (rank : 79) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.089040 (rank : 66) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.070731 (rank : 93) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.064885 (rank : 108) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.051134 (rank : 176) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA61_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.114495 (rank : 33) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI64 | Gene names | KRTAP6-1, KAP6.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-1. | |||||
|
KRA62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.087330 (rank : 69) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 2 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI66 | Gene names | KRTAP6-2, KAP6.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-2. | |||||
|
KRA63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.161735 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3LI67 | Gene names | KRTAP6-3, KAP6.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-3. | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.055700 (rank : 145) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055792 (rank : 143) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.068759 (rank : 101) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050003 (rank : 188) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050395 (rank : 185) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.052292 (rank : 170) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MAGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.056819 (rank : 139) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |
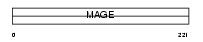
|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.053044 (rank : 166) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |
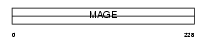
|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.053447 (rank : 161) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |
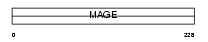
|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.055789 (rank : 144) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.052610 (rank : 167) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGA8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.055012 (rank : 148) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
MAGA9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.056791 (rank : 140) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.057552 (rank : 137) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
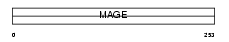
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.058310 (rank : 134) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |
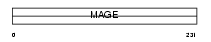
|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGAC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.053050 (rank : 165) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |
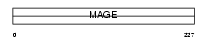
|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.059891 (rank : 122) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |
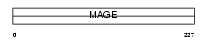
|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.061913 (rank : 118) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.063470 (rank : 113) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAGB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.062065 (rank : 117) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.065100 (rank : 107) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGBA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.063541 (rank : 112) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGBI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.059846 (rank : 124) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.058487 (rank : 131) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.058133 (rank : 135) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.053306 (rank : 162) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.094015 (rank : 60) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.103500 (rank : 50) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.092464 (rank : 64) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.093564 (rank : 62) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.084746 (rank : 70) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.069131 (rank : 100) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.072483 (rank : 90) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.070133 (rank : 95) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.069970 (rank : 96) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.073177 (rank : 88) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.073508 (rank : 85) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.071000 (rank : 92) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
MAGL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.071307 (rank : 91) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |
|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MMGL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.052572 (rank : 168) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |
|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
MN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.067325 (rank : 103) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
MORN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.121469 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
MORN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.092305 (rank : 65) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
MORN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.088728 (rank : 67) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
MORN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.127260 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.126758 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MRC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.054297 (rank : 157) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.054721 (rank : 153) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.075915 (rank : 78) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NECD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.077293 (rank : 75) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
|
|||||
| Description | Necdin. | |||||
|
NECD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.076182 (rank : 77) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.050263 (rank : 186) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUP62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.051106 (rank : 177) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
NUP62_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.059037 (rank : 129) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.097333 (rank : 55) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
NUPL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054158 (rank : 158) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BVL2, O43160 | Gene names | NUPL1, KIAA0410 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.109372 (rank : 42) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.108382 (rank : 43) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.316335 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.059201 (rank : 128) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PO121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.082978 (rank : 72) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
RBP56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.079208 (rank : 74) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
REG1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.055811 (rank : 142) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |
|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
REG3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.054311 (rank : 156) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
REG3B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.054670 (rank : 154) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
REG3G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.055455 (rank : 146) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
REG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.059866 (rank : 123) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.069897 (rank : 97) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
SIAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.054462 (rank : 155) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.063643 (rank : 111) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SRCA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.069453 (rank : 98) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.107452 (rank : 45) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.098178 (rank : 53) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
SUSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.063169 (rank : 115) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
T2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.062293 (rank : 116) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
TETN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.050402 (rank : 184) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |
|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.064761 (rank : 109) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.064278 (rank : 110) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TSGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.128854 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
TSGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.120423 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
XLKD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.073253 (rank : 87) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
XLKD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.074909 (rank : 81) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.09739e-05 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.698669 (rank : 2) | θ value | 5.44631e-05 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
JPH4_HUMAN
|
||||||
| NC score | 0.356069 (rank : 3) | θ value | 0.00102713 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.353306 (rank : 4) | θ value | 0.00102713 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.316335 (rank : 5) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
JPH3_MOUSE
|
||||||
| NC score | 0.280605 (rank : 6) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH3_HUMAN
|
||||||
| NC score | 0.280323 (rank : 7) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
KR202_HUMAN
|
||||||
| NC score | 0.275781 (rank : 8) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
KR193_HUMAN
|
||||||
| NC score | 0.256075 (rank : 9) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
JPH1_HUMAN
|
||||||
| NC score | 0.245168 (rank : 10) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.244206 (rank : 11) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.221987 (rank : 12) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.215562 (rank : 13) | θ value | 0.00102713 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.213292 (rank : 14) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.211723 (rank : 15) | θ value | 6.43352e-06 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.177554 (rank : 16) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
KR212_HUMAN
|
||||||
| NC score | 0.175201 (rank : 17) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3LI59 | Gene names | KRTAP21-2, KAP21.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-2. | |||||
|
ELN_MOUSE
|
||||||
| NC score | 0.164287 (rank : 18) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
KRA63_HUMAN
|
||||||
| NC score | 0.161735 (rank : 19) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3LI67 | Gene names | KRTAP6-3, KAP6.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-3. | |||||
|
KR197_HUMAN
|
||||||
| NC score | 0.158281 (rank : 20) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3SYF9 | Gene names | KRTAP19-7, KAP19.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-7. | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.151326 (rank : 21) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.137421 (rank : 22) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
TSGA2_HUMAN
|
||||||
| NC score | 0.128854 (rank : 23) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
MORN3_HUMAN
|
||||||
| NC score | 0.127260 (rank : 24) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN3_MOUSE
|
||||||
| NC score | 0.126758 (rank : 25) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.121469 (rank : 26) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
TSGA2_MOUSE
|
||||||
| NC score | 0.120423 (rank : 27) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.117943 (rank : 28) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
HPLN3_MOUSE
|
||||||
| NC score | 0.116408 (rank : 29) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
HPLN3_HUMAN
|
||||||
| NC score | 0.116234 (rank : 30) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
KR191_HUMAN
|
||||||
| NC score | 0.114737 (rank : 31) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUB9, Q3LI75 | Gene names | KRTAP19-1, KAP19.1, KRTAP19.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-1 (High tyrosine-glycine keratin- associated protein 19.1). | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.114513 (rank : 32) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
KRA61_HUMAN
|
||||||
| NC score | 0.114495 (rank : 33) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI64 | Gene names | KRTAP6-1, KAP6.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-1. | |||||
|
HPLN2_MOUSE
|
||||||
| NC score | 0.114010 (rank : 34) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.113458 (rank : 35) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
HPLN2_HUMAN
|
||||||
| NC score | 0.113248 (rank : 36) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.113060 (rank : 37) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
HPLN1_HUMAN
|
||||||
| NC score | 0.112898 (rank : 38) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_MOUSE
|
||||||
| NC score | 0.112004 (rank : 39) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN4_HUMAN
|
||||||
| NC score | 0.111207 (rank : 40) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
HPLN4_MOUSE
|
||||||
| NC score | 0.109409 (rank : 41) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.109372 (rank : 42) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.108382 (rank : 43) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
KR211_HUMAN
|
||||||
| NC score | 0.108264 (rank : 44) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI58 | Gene names | KRTAP21-1, KAP21.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-1. | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.107452 (rank : 45) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
ELN_HUMAN
|
||||||
| NC score | 0.107074 (rank : 46) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.106905 (rank : 47) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KR195_HUMAN
|
||||||
| NC score | 0.104589 (rank : 48) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI72 | Gene names | KRTAP19-5, KAP19.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-5. | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.104581 (rank : 49) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
MAGD1_MOUSE
|
||||||
| NC score | 0.103500 (rank : 50) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.100969 (rank : 51) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.100604 (rank : 52) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.098178 (rank : 53) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.098162 (rank : 54) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.097333 (rank : 55) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.097317 (rank : 56) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.096396 (rank : 57) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.096220 (rank : 58) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
KR131_HUMAN
|
||||||
| NC score | 0.095303 (rank : 59) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IUC0, Q3LI79 | Gene names | KRTAP13-1, KAP13.1, KRTAP13.1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin-associated protein 13-1 (High sulfur keratin-associated protein 13.1). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.094015 (rank : 60) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.093899 (rank : 61) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.093564 (rank : 62) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
KR192_HUMAN
|
||||||
| NC score | 0.093049 (rank : 63) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LHN2 | Gene names | KRTAP19-2, KAP19.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-2. | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.092464 (rank : 64) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MORN2_HUMAN
|
||||||
| NC score | 0.092305 (rank : 65) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.089040 (rank : 66) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
MORN2_MOUSE
|
||||||
| NC score | 0.088728 (rank : 67) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.088190 (rank : 68) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA62_HUMAN
|
||||||
| NC score | 0.087330 (rank : 69) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 2 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI66 | Gene names | KRTAP6-2, KAP6.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-2. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.084746 (rank : 70) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.083012 (rank : 71) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.082978 (rank : 72) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
FA98A_HUMAN
|
||||||
| NC score | 0.082231 (rank : 73) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCA5 | Gene names | FAM98A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
RBP56_HUMAN
|
||||||
| NC score | 0.079208 (rank : 74) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.077293 (rank : 75) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.076480 (rank : 76) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
NECD_MOUSE
|
||||||
| NC score | 0.076182 (rank : 77) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.075915 (rank : 78) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.075831 (rank : 79) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.075512 (rank : 80) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
XLKD1_MOUSE
|
||||||
| NC score | 0.074909 (rank : 81) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.074888 (rank : 82) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CDSN_HUMAN
|
||||||
| NC score | 0.074752 (rank : 83) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.073581 (rank : 84) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
MAGH1_MOUSE
|
||||||
| NC score | 0.073508 (rank : 85) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.073470 (rank : 86) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
XLKD1_HUMAN
|
||||||
| NC score | 0.073253 (rank : 87) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
MAGH1_HUMAN
|
||||||
| NC score | 0.073177 (rank : 88) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
FA98A_MOUSE
|
||||||
| NC score | 0.072555 (rank : 89) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TJZ6, Q99KJ2 | Gene names | Fam98a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
MAGF1_HUMAN
|
||||||
| NC score | 0.072483 (rank : 90) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGL2_MOUSE
|
||||||
| NC score | 0.071307 (rank : 91) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MAGL2_HUMAN
|
||||||
| NC score | 0.071000 (rank : 92) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.070731 (rank : 93) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.070670 (rank : 94) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
MAGG1_HUMAN
|
||||||
| NC score | 0.070133 (rank : 95) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGG1_MOUSE
|
||||||
| NC score | 0.069970 (rank : 96) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.069897 (rank : 97) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.069453 (rank : 98) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
KR132_HUMAN
|
||||||
| NC score | 0.069257 (rank : 99) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q52LG2 | Gene names | KRTAP13-2, KAP13.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-2. | |||||
|
MAGE2_HUMAN
|
||||||
| NC score | 0.069131 (rank : 100) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.068759 (rank : 101) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.067526 (rank : 102) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.067325 (rank : 103) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.066303 (rank : 104) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.065948 (rank : 105) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.065211 (rank : 106) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.065100 (rank : 107) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.064885 (rank : 108) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.064761 (rank : 109) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.064278 (rank : 110) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.063643 (rank : 111) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MAGBA_HUMAN
|
||||||
| NC score | 0.063541 (rank : 112) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGB3_HUMAN
|
||||||
| NC score | 0.063470 (rank : 113) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.063221 (rank : 114) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
SUSD1_HUMAN
|
||||||
| NC score | 0.063169 (rank : 115) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
T2_MOUSE
|
||||||
| NC score | 0.062293 (rank : 116) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
MAGB4_HUMAN
|
||||||
| NC score | 0.062065 (rank : 117) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGB2_HUMAN
|
||||||
| NC score | 0.061913 (rank : 118) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
CLC4D_HUMAN
|
||||||
| NC score | 0.061080 (rank : 119) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.060795 (rank : 120) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
CLC4D_MOUSE
|
||||||
| NC score | 0.060031 (rank : 121) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
MAGB1_HUMAN
|
||||||
| NC score | 0.059891 (rank : 122) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
REG4_MOUSE
|
||||||
| NC score | 0.059866 (rank : 123) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
MAGBI_HUMAN
|
||||||
| NC score | 0.059846 (rank : 124) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.059768 (rank : 125) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.059661 (rank : 126) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.059579 (rank : 127) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.059201 (rank : 128) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.059037 (rank : 129) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
CLC4G_HUMAN
|
||||||
| NC score | 0.058923 (rank : 130) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.058487 (rank : 131) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
GP123_HUMAN
|
||||||
| NC score | 0.058370 (rank : 132) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
FCER2_HUMAN
|
||||||
| NC score | 0.058313 (rank : 133) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
MAGAB_HUMAN
|
||||||
| NC score | 0.058310 (rank : 134) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGC2_HUMAN
|
||||||
| NC score | 0.058133 (rank : 135) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.057699 (rank : 136) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.057552 (rank : 137) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.057490 (rank : 138) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
MAGA1_HUMAN
|
||||||
| NC score | 0.056819 (rank : 139) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGA9_HUMAN
|
||||||
| NC score | 0.056791 (rank : 140) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
CLC4K_HUMAN
|
||||||
| NC score | 0.056608 (rank : 141) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
REG1A_HUMAN
|
||||||
| NC score | 0.055811 (rank : 142) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.055792 (rank : 143) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
MAGA4_HUMAN
|
||||||
| NC score | 0.055789 (rank : 144) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.055700 (rank : 145) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
REG3G_MOUSE
|
||||||
| NC score | 0.055455 (rank : 146) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.055121 (rank : 147) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
MAGA8_HUMAN
|
||||||
| NC score | 0.055012 (rank : 148) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.054957 (rank : 149) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FCER2_MOUSE
|
||||||
| NC score | 0.054860 (rank : 150) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
KRA13_HUMAN
|
||||||
| NC score | 0.054775 (rank : 151) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
CLC4F_MOUSE
|
||||||
| NC score | 0.054757 (rank : 152) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.054721 (rank : 153) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
REG3B_MOUSE
|
||||||
| NC score | 0.054670 (rank : 154) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
SIAL_HUMAN
|
||||||
| NC score | 0.054462 (rank : 155) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
REG3A_HUMAN
|
||||||
| NC score | 0.054311 (rank : 156) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.054297 (rank : 157) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
NUPL1_HUMAN
|
||||||
| NC score | 0.054158 (rank : 158) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BVL2, O43160 | Gene names | NUPL1, KIAA0410 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.053832 (rank : 159) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
CLC4G_MOUSE
|
||||||
| NC score | 0.053607 (rank : 160) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
MAGA3_HUMAN
|
||||||
| NC score | 0.053447 (rank : 161) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGC3_HUMAN
|
||||||
| NC score | 0.053306 (rank : 162) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.053255 (rank : 163) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
CLC4M_HUMAN
|
||||||
| NC score | 0.053060 (rank : 164) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
MAGAC_HUMAN
|
||||||
| NC score | 0.053050 (rank : 165) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA2_HUMAN
|
||||||
| NC score | 0.053044 (rank : 166) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGA6_HUMAN
|
||||||
| NC score | 0.052610 (rank : 167) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MMGL_MOUSE
|
||||||
| NC score | 0.052572 (rank : 168) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
C209C_MOUSE
|
||||||
| NC score | 0.052432 (rank : 169) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.052292 (rank : 170) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
CD209_HUMAN
|
||||||
| NC score | 0.052146 (rank : 171) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
ASGR2_MOUSE
|
||||||
| NC score | 0.051914 (rank : 172) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
CLC4K_MOUSE
|
||||||
| NC score | 0.051521 (rank : 173) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.051456 (rank : 174) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
CLC4E_MOUSE
|
||||||
| NC score | 0.051232 (rank : 175) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.051134 (rank : 176) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
NUP62_HUMAN
|
||||||
| NC score | 0.051106 (rank : 177) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
EGFL7_MOUSE
|
||||||
| NC score | 0.050891 (rank : 178) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXT5, Q6XD35, Q9DCP5 | Gene names | Egfl7, Megf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 7 precursor (Multiple EGF-like domain protein 7) (Multiple epidermal growth factor-like domain protein 7) (Vascular endothelial statin) (VE-statin) (NOTCH4-like protein) (Zneu1). | |||||
|
ASGR1_HUMAN
|
||||||
| NC score | 0.050785 (rank : 179) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07306 | Gene names | ASGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin H1). | |||||
|
K1C9_HUMAN
|
||||||
| NC score | 0.050740 (rank : 180) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
CLC10_HUMAN
|
||||||
| NC score | 0.050683 (rank : 181) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.050510 (rank : 182) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
CD302_MOUSE
|
||||||
| NC score | 0.050421 (rank : 183) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
TETN_HUMAN
|
||||||
| NC score | 0.050402 (rank : 184) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |

|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.050395 (rank : 185) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.050263 (rank : 186) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.050152 (rank : 187) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.050003 (rank : 188) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.047626 (rank : 189) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
KRHB1_HUMAN
|
||||||
| NC score | 0.030362 (rank : 190) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB3_HUMAN
|
||||||
| NC score | 0.026577 (rank : 191) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.025862 (rank : 192) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
ZN297_MOUSE
|
||||||
| NC score | 0.019192 (rank : 193) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0G7 | Gene names | Znf297, Bing1, Zfp297 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 297 (BING1 protein). | |||||
|
FOXF1_HUMAN
|
||||||
| NC score | 0.009840 (rank : 194) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12946, Q5FWE5 | Gene names | FOXF1, FKHL5, FREAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Forkhead-related activator 1). | |||||