Please be patient as the page loads
|
MORN1_HUMAN
|
||||||
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MORN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
MORN3_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 2) | NC score | 0.791240 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN3_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 3) | NC score | 0.786212 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
TSGA2_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 4) | NC score | 0.795458 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
TSGA2_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 5) | NC score | 0.794186 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 6) | NC score | 0.522228 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 7) | NC score | 0.522197 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
JPH3_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 8) | NC score | 0.638219 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH3_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 9) | NC score | 0.636807 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 10) | NC score | 0.624134 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 11) | NC score | 0.622742 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 12) | NC score | 0.583660 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 13) | NC score | 0.609764 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH4_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 14) | NC score | 0.610860 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 15) | NC score | 0.606660 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
SETD7_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 16) | NC score | 0.492924 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
SETD7_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 17) | NC score | 0.495833 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 18) | NC score | 0.349036 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
MORN2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 19) | NC score | 0.621420 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
MORN2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 20) | NC score | 0.599610 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.027367 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
CO4A4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.046559 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
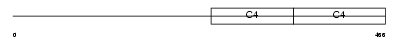
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.058549 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.049345 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.050700 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.063645 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.030254 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.043308 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
HXD11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.021942 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31277, Q9NS02 | Gene names | HOXD11, HOX4F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4F). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.044917 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MYPN_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.020647 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.029854 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.053836 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.043513 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.043531 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
SYT3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.020557 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.037771 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.036912 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PBX2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.032127 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P40425 | Gene names | PBX2, G17 | |||
|
Domain Architecture |
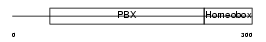
|
|||||
| Description | Pre-B-cell leukemia transcription factor 2 (Homeobox protein PBX2) (Protein G17). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.043539 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
HXD11_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.019362 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23813 | Gene names | Hoxd11, Hox-4.6, Hoxd-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4.6) (Hox-5.5). | |||||
|
PBX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.028833 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35984 | Gene names | Pbx2 | |||
|
Domain Architecture |
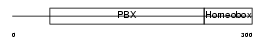
|
|||||
| Description | Pre-B-cell leukemia transcription factor 2 (Homeobox protein PBX2). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.038270 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SYT3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.018956 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.026131 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.015850 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
CT151_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.033528 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
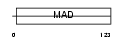
|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
HDAC7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.015352 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUI4, Q7Z5I1, Q96K01, Q9BR73, Q9H7L0, Q9NW41, Q9NWA9, Q9NYK9, Q9UFU7 | Gene names | HDAC7A, HDAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.012157 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
BCL7C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.027747 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.035742 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.020252 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.013269 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.017373 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.016886 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.032633 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.032633 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.040838 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ZN628_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | -0.001931 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.042480 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.031849 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
EVI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | -0.001156 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14404 | Gene names | Evi1, Evi-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
HXA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.007174 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.006322 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.019019 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
VASP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.021781 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
CMTA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.014379 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.021943 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LAGE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.026103 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14657, Q5HY39, Q8IZ78 | Gene names | LAGE3, DXS9879E, ESO3, ITBA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L antigen family member 3 (Protein ITBA2) (Protein ESO-3). | |||||
|
M3K1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | -0.002101 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53349, Q60831, Q9R0U3, Q9R256 | Gene names | Map3k1, Mekk, Mekk1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 1 (EC 2.7.11.25) (MAPK/ERK kinase kinase 1) (MEK kinase 1) (MEKK 1). | |||||
|
MBD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.011999 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBB5, O95242, Q9UIS8 | Gene names | MBD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2) (Demethylase) (DMTase). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.024613 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.026863 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TULP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.036797 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
CAPS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.008677 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
CENB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.011634 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.037928 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.028401 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.012536 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.009555 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
IRX3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.010395 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
NBL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.015508 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61477 | Gene names | Nbl1, Dan, Dana | |||
|
Domain Architecture |
|
|||||
| Description | Neuroblastoma suppressor of tumorigenicity 1 precursor (Zinc finger protein DAN) (N03). | |||||
|
P85B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.004809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.002920 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.031369 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.053793 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.009862 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TF3C2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.012405 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BL74, Q3U7Z9, Q80XQ7, Q8BJJ4, Q8BLP3, Q8K1J1, Q9CSS0, Q9CVE6, Q9DBK6 | Gene names | Gtf3c2, Kiaa0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.001510 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
KR193_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.074954 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
KR202_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.092176 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.121469 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.113852 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
TSGA2_HUMAN
|
||||||
| NC score | 0.795458 (rank : 2) | θ value | 1.47631e-18 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
TSGA2_MOUSE
|
||||||
| NC score | 0.794186 (rank : 3) | θ value | 2.5182e-18 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
MORN3_MOUSE
|
||||||
| NC score | 0.791240 (rank : 4) | θ value | 1.74391e-19 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN3_HUMAN
|
||||||
| NC score | 0.786212 (rank : 5) | θ value | 8.65492e-19 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
JPH3_HUMAN
|
||||||
| NC score | 0.638219 (rank : 6) | θ value | 2.0648e-12 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH3_MOUSE
|
||||||
| NC score | 0.636807 (rank : 7) | θ value | 4.59992e-12 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH1_HUMAN
|
||||||
| NC score | 0.624134 (rank : 8) | θ value | 6.64225e-11 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.622742 (rank : 9) | θ value | 6.64225e-11 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
MORN2_HUMAN
|
||||||
| NC score | 0.621420 (rank : 10) | θ value | 8.40245e-06 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
JPH4_HUMAN
|
||||||
| NC score | 0.610860 (rank : 11) | θ value | 3.64472e-09 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.609764 (rank : 12) | θ value | 1.47974e-10 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.606660 (rank : 13) | θ value | 3.64472e-09 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
MORN2_MOUSE
|
||||||
| NC score | 0.599610 (rank : 14) | θ value | 2.44474e-05 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.583660 (rank : 15) | θ value | 8.67504e-11 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.522228 (rank : 16) | θ value | 3.40345e-15 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.522197 (rank : 17) | θ value | 3.40345e-15 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
SETD7_HUMAN
|
||||||
| NC score | 0.495833 (rank : 18) | θ value | 4.76016e-09 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
SETD7_MOUSE
|
||||||
| NC score | 0.492924 (rank : 19) | θ value | 3.64472e-09 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.349036 (rank : 20) | θ value | 3.41135e-07 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.121469 (rank : 21) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.113852 (rank : 22) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
KR202_HUMAN
|
||||||
| NC score | 0.092176 (rank : 23) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
KR193_HUMAN
|
||||||
| NC score | 0.074954 (rank : 24) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.063645 (rank : 25) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.058549 (rank : 26) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.053836 (rank : 27) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.053793 (rank : 28) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.050700 (rank : 29) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.049345 (rank : 30) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.046559 (rank : 31) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.044917 (rank : 32) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.043539 (rank : 33) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.043531 (rank : 34) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.043513 (rank : 35) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.043308 (rank : 36) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.042480 (rank : 37) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.040838 (rank : 38) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.038270 (rank : 39) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.037928 (rank : 40) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.037771 (rank : 41) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.036912 (rank : 42) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TULP4_MOUSE
|
||||||
| NC score | 0.036797 (rank : 43) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.035742 (rank : 44) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.033528 (rank : 45) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.032633 (rank : 46) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.032633 (rank : 47) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
PBX2_HUMAN
|
||||||
| NC score | 0.032127 (rank : 48) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P40425 | Gene names | PBX2, G17 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-B-cell leukemia transcription factor 2 (Homeobox protein PBX2) (Protein G17). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.031849 (rank : 49) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.031369 (rank : 50) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.030254 (rank : 51) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.029854 (rank : 52) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
PBX2_MOUSE
|
||||||
| NC score | 0.028833 (rank : 53) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35984 | Gene names | Pbx2 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-B-cell leukemia transcription factor 2 (Homeobox protein PBX2). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.028401 (rank : 54) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
BCL7C_HUMAN
|
||||||
| NC score | 0.027747 (rank : 55) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.027367 (rank : 56) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.026863 (rank : 57) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.026131 (rank : 58) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
LAGE3_HUMAN
|
||||||
| NC score | 0.026103 (rank : 59) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14657, Q5HY39, Q8IZ78 | Gene names | LAGE3, DXS9879E, ESO3, ITBA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L antigen family member 3 (Protein ITBA2) (Protein ESO-3). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.024613 (rank : 60) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.021943 (rank : 61) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HXD11_HUMAN
|
||||||
| NC score | 0.021942 (rank : 62) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31277, Q9NS02 | Gene names | HOXD11, HOX4F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4F). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.021781 (rank : 63) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
MYPN_MOUSE
|
||||||
| NC score | 0.020647 (rank : 64) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.020557 (rank : 65) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.020252 (rank : 66) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
HXD11_MOUSE
|
||||||
| NC score | 0.019362 (rank : 67) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23813 | Gene names | Hoxd11, Hox-4.6, Hoxd-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4.6) (Hox-5.5). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.019019 (rank : 68) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.018956 (rank : 69) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.017373 (rank : 70) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.016886 (rank : 71) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.015850 (rank : 72) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
NBL1_MOUSE
|
||||||
| NC score | 0.015508 (rank : 73) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61477 | Gene names | Nbl1, Dan, Dana | |||
|
Domain Architecture |

|
|||||
| Description | Neuroblastoma suppressor of tumorigenicity 1 precursor (Zinc finger protein DAN) (N03). | |||||
|
HDAC7_HUMAN
|
||||||
| NC score | 0.015352 (rank : 74) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUI4, Q7Z5I1, Q96K01, Q9BR73, Q9H7L0, Q9NW41, Q9NWA9, Q9NYK9, Q9UFU7 | Gene names | HDAC7A, HDAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
CMTA2_HUMAN
|
||||||
| NC score | 0.014379 (rank : 75) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.013269 (rank : 76) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.012536 (rank : 77) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
TF3C2_MOUSE
|
||||||
| NC score | 0.012405 (rank : 78) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BL74, Q3U7Z9, Q80XQ7, Q8BJJ4, Q8BLP3, Q8K1J1, Q9CSS0, Q9CVE6, Q9DBK6 | Gene names | Gtf3c2, Kiaa0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.012157 (rank : 79) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
MBD2_HUMAN
|
||||||
| NC score | 0.011999 (rank : 80) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBB5, O95242, Q9UIS8 | Gene names | MBD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2) (Demethylase) (DMTase). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.011634 (rank : 81) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
IRX3_HUMAN
|
||||||
| NC score | 0.010395 (rank : 82) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.009862 (rank : 83) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.009555 (rank : 84) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
CAPS2_MOUSE
|
||||||
| NC score | 0.008677 (rank : 85) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
HXA4_MOUSE
|
||||||
| NC score | 0.007174 (rank : 86) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.006322 (rank : 87) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.004809 (rank : 88) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.002920 (rank : 89) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.001510 (rank : 90) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
EVI1_MOUSE
|
||||||
| NC score | -0.001156 (rank : 91) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14404 | Gene names | Evi1, Evi-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
ZN628_HUMAN
|
||||||
| NC score | -0.001931 (rank : 92) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
M3K1_MOUSE
|
||||||
| NC score | -0.002101 (rank : 93) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53349, Q60831, Q9R0U3, Q9R256 | Gene names | Map3k1, Mekk, Mekk1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 1 (EC 2.7.11.25) (MAPK/ERK kinase kinase 1) (MEK kinase 1) (MEKK 1). | |||||