Please be patient as the page loads
|
CLIC6_MOUSE
|
||||||
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CLIC6_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 1.12192e-159 (rank : 2) | NC score | 0.902385 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CLIC4_HUMAN
|
||||||
| θ value | 7.14227e-106 (rank : 3) | NC score | 0.958566 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y696, Q9UFW9, Q9UQJ6 | Gene names | CLIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (Intracellular chloride ion channel protein p64H1). | |||||
|
CLIC4_MOUSE
|
||||||
| θ value | 2.71407e-105 (rank : 4) | NC score | 0.958478 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYB1, Q8BMG5 | Gene names | Clic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (mc3s5/mtCLIC). | |||||
|
CLIC5_MOUSE
|
||||||
| θ value | 7.39111e-103 (rank : 5) | NC score | 0.958295 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BXK9, Q3U0H8 | Gene names | Clic5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
CLIC5_HUMAN
|
||||||
| θ value | 2.15048e-102 (rank : 6) | NC score | 0.953452 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZA1, Q5T4Z0, Q96JT5, Q9BWZ0 | Gene names | CLIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
CLIC2_HUMAN
|
||||||
| θ value | 1.30768e-91 (rank : 7) | NC score | 0.957445 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15247, O15174, Q5JT80, Q8TCE3 | Gene names | CLIC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 2 (XAP121). | |||||
|
CLIC1_HUMAN
|
||||||
| θ value | 3.11973e-85 (rank : 8) | NC score | 0.954413 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00299, Q15089 | Gene names | CLIC1, NCC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27) (Chloride channel ABP) (Regulatory nuclear chloride ion channel protein) (hRNCC). | |||||
|
CLIC1_MOUSE
|
||||||
| θ value | 1.54831e-84 (rank : 9) | NC score | 0.954222 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1Q5 | Gene names | Clic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27). | |||||
|
CLIC3_HUMAN
|
||||||
| θ value | 1.55911e-60 (rank : 10) | NC score | 0.940201 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95833 | Gene names | CLIC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 3. | |||||
|
CLIC3_MOUSE
|
||||||
| θ value | 3.84025e-59 (rank : 11) | NC score | 0.937808 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7P7 | Gene names | Clic3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 3. | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 12) | NC score | 0.059862 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 13) | NC score | 0.059876 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 14) | NC score | 0.071114 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 15) | NC score | 0.087121 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 16) | NC score | 0.072110 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ASPX_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 17) | NC score | 0.117181 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 18) | NC score | 0.043135 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 19) | NC score | 0.064921 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
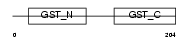
GSTO1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 20) | NC score | 0.277717 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O09131, Q3TH87 | Gene names | Gsto1, Gstx, Gtsttl | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1) (p28). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 21) | NC score | 0.046661 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 22) | NC score | 0.047061 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 23) | NC score | 0.011296 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 24) | NC score | 0.042206 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PASK_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 25) | NC score | 0.008525 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 26) | NC score | 0.108057 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.067634 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.027172 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.041372 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.025959 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.023583 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
GBX2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.009693 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48031 | Gene names | Gbx2, Mmoxa, Stra7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2) (Homeobox protein STRA7). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.060448 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.074157 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.047268 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.037108 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.076194 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.027458 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.053117 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.034420 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
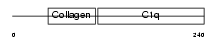
ADIPO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.018189 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60994, Q62400, Q9DC68 | Gene names | Adipoq, Acdc, Acrp30, Apm1 | |||
|
Domain Architecture |
|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipocyte-specific protein AdipoQ). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.051666 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.037172 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
ZKSC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.002785 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.041272 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.045704 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.039808 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.083077 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.037344 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.078353 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TRI56_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.019447 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.033740 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
CF150_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.042986 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.047326 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.039377 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.032723 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.036546 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.038424 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.043738 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
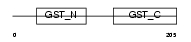
GSTO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.170775 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78417, Q5TA03, Q7Z3T2 | Gene names | GSTO1, GSTTLP28 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1). | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.041158 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.012233 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.023486 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.007397 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.026420 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.024880 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
GDAP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.063031 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TB36 | Gene names | GDAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
IRAK4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.002588 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.004763 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.024396 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
RA54B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.011438 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.021878 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.016542 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
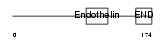
EDN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.022857 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14138, Q03229 | Gene names | EDN3 | |||
|
Domain Architecture |
|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.032950 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.027058 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.038718 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.024046 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.122606 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TAU_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.050493 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.025007 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
CCDC8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.087828 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0W5, Q8TB26 | Gene names | CCDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 8. | |||||
|
CDKL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.001366 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUK0, Q3UL60, Q3V3X7, Q9QYI1, Q9QYI2 | Gene names | Cdkl2, Kkiamre, Kkm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE). | |||||
|
MAAI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.078438 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVL0 | Gene names | Gstz1, Maai | |||
|
Domain Architecture |
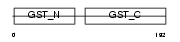
|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
NRK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.006416 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
PPT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.016983 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50897 | Gene names | PPT1, PPT | |||
|
Domain Architecture |
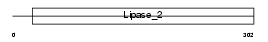
|
|||||
| Description | Palmitoyl-protein thioesterase 1 precursor (EC 3.1.2.22) (PPT-1) (Palmitoyl-protein hydrolase 1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.057875 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.027768 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.001079 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
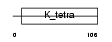
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
HDAC6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.009683 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
IP3KC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.011144 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
MORN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.013269 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.020710 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.028826 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.015408 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.010281 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
AATK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.004938 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.060779 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
BSCL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.016287 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.028521 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.069209 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.005503 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.036429 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.014171 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
TRI29_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.007785 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
ZN432_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | -0.000990 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94892 | Gene names | ZNF432, KIAA0798 | |||
|
Domain Architecture |
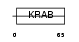
|
|||||
| Description | Zinc finger protein 432. | |||||
|
ZN467_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | -0.000809 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8JZL0, Q9JJ98 | Gene names | Znf467, Ezi, Zfp467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467 (Endothelial cell-derived zinc finger protein) (EZI). | |||||
|
ZSCA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.000329 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NAM6 | Gene names | ZSCAN4, ZNF494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 4 (Zinc finger protein 494). | |||||
|
BCAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.023723 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.039557 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.019412 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.074836 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
ES8L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.005464 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.025543 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GAGE6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.009865 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13070 | Gene names | GAGE6 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen 6 (GAGE-6). | |||||
|
GDAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.062001 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88741, Q8C7Q5, Q9CTN2 | Gene names | Gdap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
GOPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.005089 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
GP101_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | -0.002158 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96P66, Q8NG93 | Gene names | GPR101 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 101. | |||||
|
GSTO2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.074496 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K2Q2 | Gene names | Gsto2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutathione transferase omega-2 (EC 2.5.1.18). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.043687 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.029137 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.011116 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
S12A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.007905 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
TSKS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.012146 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.002442 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.016784 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
GAGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.013490 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75459, Q6FGM3, Q9BSS7 | Gene names | PAGE1, GAGE9, GAGEB1 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen family B member 1 (Prostate-associated gene 1 protein) (PAGE-1) (GAGE-9) (AL5). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.003258 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
PANK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.005989 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.023150 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TMED8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.009549 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PL24, Q9P1V9 | Gene names | TMED8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TMED8. | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.026546 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ZN174_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | -0.000226 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.000838 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.105680 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.122953 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.050835 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.052200 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.059634 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.064478 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
MAAI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.069871 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43708, O15308, O75430, Q7Z610, Q9BV63 | Gene names | GSTZ1, MAAI | |||
|
Domain Architecture |
|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.129372 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.126775 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.072709 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
MRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.069494 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.058687 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.072395 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.057994 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.059038 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.067400 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.072494 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.055901 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.064571 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.064975 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.055458 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.098500 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.052617 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.064405 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.068128 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.075644 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.052815 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.059177 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.056350 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CLIC4_HUMAN
|
||||||
| NC score | 0.958566 (rank : 2) | θ value | 7.14227e-106 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y696, Q9UFW9, Q9UQJ6 | Gene names | CLIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (Intracellular chloride ion channel protein p64H1). | |||||
|
CLIC4_MOUSE
|
||||||
| NC score | 0.958478 (rank : 3) | θ value | 2.71407e-105 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYB1, Q8BMG5 | Gene names | Clic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (mc3s5/mtCLIC). | |||||
|
CLIC5_MOUSE
|
||||||
| NC score | 0.958295 (rank : 4) | θ value | 7.39111e-103 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BXK9, Q3U0H8 | Gene names | Clic5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
CLIC2_HUMAN
|
||||||
| NC score | 0.957445 (rank : 5) | θ value | 1.30768e-91 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15247, O15174, Q5JT80, Q8TCE3 | Gene names | CLIC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 2 (XAP121). | |||||
|
CLIC1_HUMAN
|
||||||
| NC score | 0.954413 (rank : 6) | θ value | 3.11973e-85 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00299, Q15089 | Gene names | CLIC1, NCC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27) (Chloride channel ABP) (Regulatory nuclear chloride ion channel protein) (hRNCC). | |||||
|
CLIC1_MOUSE
|
||||||
| NC score | 0.954222 (rank : 7) | θ value | 1.54831e-84 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1Q5 | Gene names | Clic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27). | |||||
|
CLIC5_HUMAN
|
||||||
| NC score | 0.953452 (rank : 8) | θ value | 2.15048e-102 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZA1, Q5T4Z0, Q96JT5, Q9BWZ0 | Gene names | CLIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
CLIC3_HUMAN
|
||||||
| NC score | 0.940201 (rank : 9) | θ value | 1.55911e-60 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95833 | Gene names | CLIC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 3. | |||||
|
CLIC3_MOUSE
|
||||||
| NC score | 0.937808 (rank : 10) | θ value | 3.84025e-59 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7P7 | Gene names | Clic3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 3. | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.902385 (rank : 11) | θ value | 1.12192e-159 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
GSTO1_MOUSE
|
||||||
| NC score | 0.277717 (rank : 12) | θ value | 0.00175202 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O09131, Q3TH87 | Gene names | Gsto1, Gstx, Gtsttl | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1) (p28). | |||||
|
GSTO1_HUMAN
|
||||||
| NC score | 0.170775 (rank : 13) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78417, Q5TA03, Q7Z3T2 | Gene names | GSTO1, GSTTLP28 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.129372 (rank : 14) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.126775 (rank : 15) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.122953 (rank : 16) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.122606 (rank : 17) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.117181 (rank : 18) | θ value | 0.00102713 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.108057 (rank : 19) | θ value | 0.0148317 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.105680 (rank : 20) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.098500 (rank : 21) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CCDC8_HUMAN
|
||||||
| NC score | 0.087828 (rank : 22) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0W5, Q8TB26 | Gene names | CCDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 8. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.087121 (rank : 23) | θ value | 7.1131e-05 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.083077 (rank : 24) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MAAI_MOUSE
|
||||||
| NC score | 0.078438 (rank : 25) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVL0 | Gene names | Gstz1, Maai | |||
|
Domain Architecture |

|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.078353 (rank : 26) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.076194 (rank : 27) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.075644 (rank : 28) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.074836 (rank : 29) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GSTO2_MOUSE
|
||||||
| NC score | 0.074496 (rank : 30) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K2Q2 | Gene names | Gsto2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutathione transferase omega-2 (EC 2.5.1.18). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.074157 (rank : 31) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.072709 (rank : 32) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.072494 (rank : 33) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.072395 (rank : 34) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.072110 (rank : 35) | θ value | 0.000786445 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.071114 (rank : 36) | θ value | 3.41135e-07 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
MAAI_HUMAN
|
||||||
| NC score | 0.069871 (rank : 37) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43708, O15308, O75430, Q7Z610, Q9BV63 | Gene names | GSTZ1, MAAI | |||
|
Domain Architecture |

|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.069494 (rank : 38) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.069209 (rank : 39) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.068128 (rank : 40) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.067634 (rank : 41) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.067400 (rank : 42) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.064975 (rank : 43) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.064921 (rank : 44) | θ value | 0.00134147 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.064571 (rank : 45) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.064478 (rank : 46) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.064405 (rank : 47) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
GDAP1_HUMAN
|
||||||
| NC score | 0.063031 (rank : 48) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TB36 | Gene names | GDAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
GDAP1_MOUSE
|
||||||
| NC score | 0.062001 (rank : 49) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88741, Q8C7Q5, Q9CTN2 | Gene names | Gdap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.060779 (rank : 50) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.060448 (rank : 51) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.059876 (rank : 52) | θ value | 3.41135e-07 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.059862 (rank : 53) | θ value | 8.97725e-08 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.059634 (rank : 54) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.059177 (rank : 55) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.059038 (rank : 56) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.058687 (rank : 57) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.057994 (rank : 58) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.057875 (rank : 59) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.056350 (rank : 60) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.055901 (rank : 61) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.055458 (rank : 62) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.053117 (rank : 63) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.052815 (rank : 64) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.052617 (rank : 65) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.052200 (rank : 66) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.051666 (rank : 67) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.050835 (rank : 68) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.050493 (rank : 69) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.047326 (rank : 70) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.047268 (rank : 71) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.047061 (rank : 72) | θ value | 0.00509761 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.046661 (rank : 73) | θ value | 0.00298849 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.045704 (rank : 74) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.043738 (rank : 75) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.043687 (rank : 76) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.043135 (rank : 77) | θ value | 0.00102713 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CF150_HUMAN
|
||||||
| NC score | 0.042986 (rank : 78) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N884, Q32NC9, Q5SWL0, Q5SWL1, Q96E45 | Gene names | C6orf150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf150. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.042206 (rank : 79) | θ value | 0.0113563 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.041372 (rank : 80) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.041272 (rank : 81) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.041158 (rank : 82) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.039808 (rank : 83) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.039557 (rank : 84) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.039377 (rank : 85) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.038718 (rank : 86) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.038424 (rank : 87) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.037344 (rank : 88) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.037172 (rank : 89) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.037108 (rank : 90) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.036546 (rank : 91) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.036429 (rank : 92) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.034420 (rank : 93) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.033740 (rank : 94) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.032950 (rank : 95) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.032723 (rank : 96) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.029137 (rank : 97) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.028826 (rank : 98) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.028521 (rank : 99) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.027768 (rank : 100) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.027458 (rank : 101) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.027172 (rank : 102) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.027058 (rank : 103) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.026546 (rank : 104) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.026420 (rank : 105) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.025959 (rank : 106) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.025543 (rank : 107) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.025007 (rank : 108) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.024880 (rank : 109) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.024396 (rank : 110) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.024046 (rank : 111) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
BCAS1_HUMAN
|
||||||
| NC score | 0.023723 (rank : 112) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.023583 (rank : 113) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.023486 (rank : 114) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.023150 (rank : 115) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
EDN3_HUMAN
|
||||||
| NC score | 0.022857 (rank : 116) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14138, Q03229 | Gene names | EDN3 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.021878 (rank : 117) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.020710 (rank : 118) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.019447 (rank : 119) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.019412 (rank : 120) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
ADIPO_MOUSE
|
||||||
| NC score | 0.018189 (rank : 121) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60994, Q62400, Q9DC68 | Gene names | Adipoq, Acdc, Acrp30, Apm1 | |||
|
Domain Architecture |

|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipocyte-specific protein AdipoQ). | |||||
|
PPT1_HUMAN
|
||||||
| NC score | 0.016983 (rank : 122) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50897 | Gene names | PPT1, PPT | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyl-protein thioesterase 1 precursor (EC 3.1.2.22) (PPT-1) (Palmitoyl-protein hydrolase 1). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.016784 (rank : 123) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.016542 (rank : 124) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
BSCL2_MOUSE
|
||||||
| NC score | 0.016287 (rank : 125) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.015408 (rank : 126) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.014171 (rank : 127) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
GAGB1_HUMAN
|
||||||
| NC score | 0.013490 (rank : 128) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75459, Q6FGM3, Q9BSS7 | Gene names | PAGE1, GAGE9, GAGEB1 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen family B member 1 (Prostate-associated gene 1 protein) (PAGE-1) (GAGE-9) (AL5). | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.013269 (rank : 129) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.012233 (rank : 130) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
TSKS_HUMAN
|
||||||
| NC score | 0.012146 (rank : 131) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
RA54B_MOUSE
|
||||||
| NC score | 0.011438 (rank : 132) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.011296 (rank : 133) | θ value | 0.00869519 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
IP3KC_MOUSE
|
||||||
| NC score | 0.011144 (rank : 134) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.011116 (rank : 135) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.010281 (rank : 136) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
GAGE6_HUMAN
|
||||||
| NC score | 0.009865 (rank : 137) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13070 | Gene names | GAGE6 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen 6 (GAGE-6). | |||||
|
GBX2_MOUSE
|
||||||
| NC score | 0.009693 (rank : 138) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48031 | Gene names | Gbx2, Mmoxa, Stra7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2) (Homeobox protein STRA7). | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.009683 (rank : 139) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
TMED8_HUMAN
|
||||||
| NC score | 0.009549 (rank : 140) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PL24, Q9P1V9 | Gene names | TMED8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TMED8. | |||||
|
PASK_HUMAN
|
||||||
| NC score | 0.008525 (rank : 141) | θ value | 0.0113563 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||
|
S12A2_HUMAN
|
||||||
| NC score | 0.007905 (rank : 142) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
TRI29_HUMAN
|
||||||
| NC score | 0.007785 (rank : 143) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.007397 (rank : 144) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
NRK_HUMAN
|
||||||
| NC score | 0.006416 (rank : 145) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
PANK1_MOUSE
|
||||||
| NC score | 0.005989 (rank : 146) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.005503 (rank : 147) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
ES8L1_MOUSE
|
||||||
| NC score | 0.005464 (rank : 148) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
GOPC_HUMAN
|
||||||
| NC score | 0.005089 (rank : 149) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.004938 (rank : 150) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.004763 (rank : 151) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.003258 (rank : 152) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
ZKSC1_HUMAN
|
||||||
| NC score | 0.002785 (rank : 153) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||
|
IRAK4_MOUSE
|
||||||
| NC score | 0.002588 (rank : 154) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.002442 (rank : 155) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
CDKL2_MOUSE
|
||||||
| NC score | 0.001366 (rank : 156) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUK0, Q3UL60, Q3V3X7, Q9QYI1, Q9QYI2 | Gene names | Cdkl2, Kkiamre, Kkm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | 0.001079 (rank : 157) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | 0.000838 (rank : 158) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
ZSCA4_HUMAN
|
||||||
| NC score | 0.000329 (rank : 159) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NAM6 | Gene names | ZSCAN4, ZNF494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 4 (Zinc finger protein 494). | |||||
|
ZN174_HUMAN
|
||||||
| NC score | -0.000226 (rank : 160) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
ZN467_MOUSE
|
||||||
| NC score | -0.000809 (rank : 161) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8JZL0, Q9JJ98 | Gene names | Znf467, Ezi, Zfp467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467 (Endothelial cell-derived zinc finger protein) (EZI). | |||||
|
ZN432_HUMAN
|
||||||
| NC score | -0.000990 (rank : 162) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94892 | Gene names | ZNF432, KIAA0798 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 432. | |||||
|
GP101_HUMAN
|
||||||
| NC score | -0.002158 (rank : 163) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96P66, Q8NG93 | Gene names | GPR101 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 101. | |||||