Please be patient as the page loads
|
GASP2_HUMAN
|
||||||
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GASP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.946010 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP1_HUMAN
|
||||||
| θ value | 3.97187e-173 (rank : 3) | NC score | 0.934740 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP1_MOUSE
|
||||||
| θ value | 2.36115e-125 (rank : 4) | NC score | 0.902689 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
ARMX5_HUMAN
|
||||||
| θ value | 2.42216e-37 (rank : 5) | NC score | 0.833050 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1M9, Q68DB4, Q9BVZ3, Q9H969 | Gene names | ARMCX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX5_MOUSE
|
||||||
| θ value | 3.61944e-33 (rank : 6) | NC score | 0.817980 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UZB0 | Gene names | Armcx5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 7) | NC score | 0.764895 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
ARMX3_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 8) | NC score | 0.750388 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX3_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 9) | NC score | 0.749073 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 10) | NC score | 0.759135 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 11) | NC score | 0.703915 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 12) | NC score | 0.661993 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 13) | NC score | 0.101119 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ARMX6_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 14) | NC score | 0.523507 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7L4S7, Q9NWJ3 | Gene names | ARMCX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 15) | NC score | 0.110653 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 16) | NC score | 0.143999 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
ARMX4_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 17) | NC score | 0.442717 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9R4, Q5H9K8, Q8N8D6 | Gene names | ARMCX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 4. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 18) | NC score | 0.104662 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
ARMX6_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 19) | NC score | 0.476342 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3A6, Q3TUH4 | Gene names | Armcx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 20) | NC score | 0.046074 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 21) | NC score | 0.089482 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
NUFP1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 22) | NC score | 0.096994 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 23) | NC score | 0.071484 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
FUSIP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 24) | NC score | 0.100838 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 25) | NC score | 0.100838 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
K1196_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 26) | NC score | 0.047461 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
STRAD_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.077055 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.100785 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 29) | NC score | 0.116713 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.086055 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.047261 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.059755 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
YIPF2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.061639 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LP8, Q99LF3 | Gene names | Yipf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF2 (YIP1 family member 2). | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.031007 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.008263 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.037108 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.034688 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.009915 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.103939 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.071477 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.071477 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.017672 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.047140 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.013863 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.048874 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.035959 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
GNAS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.041411 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.043219 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.047114 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.070973 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.072683 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.018841 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.011813 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.047993 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
PAX1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.014954 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15863, Q642X9, Q6NTC0, Q9Y558 | Gene names | PAX1, HUP48 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-1 (HUP48). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.086225 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
M3K4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.004164 (rank : 140) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6R4, Q92612 | Gene names | MAP3K4, KIAA0213, MAPKKK4, MEKK4, MTK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4) (MAP three kinase 1). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.023966 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
PHF8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.018679 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
XPC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.033333 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01831, Q96AX0 | Gene names | XPC, XPCC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells (Xeroderma pigmentosum group C-complementing protein) (p125). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.011595 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.028366 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
GP152_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.025457 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
LC7L2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.030329 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.030660 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.017944 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.015368 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
CC054_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.027164 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX62, Q80WB2, Q8BPE9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf54 homolog. | |||||
|
ERCC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.030452 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
LGI3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.010748 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N145, Q86TL4, Q8N296 | Gene names | LGI3, LGIL4 | |||
|
Domain Architecture |
|
|||||
| Description | Leucine-rich repeat LGI family member 3 precursor (Leucine-rich glioma-inactivated protein 3) (LGI1-like protein 4). | |||||
|
LGI3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.010836 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K406 | Gene names | Lgi3 | |||
|
Domain Architecture |
|
|||||
| Description | Leucine-rich repeat LGI family member 3 precursor (Leucine-rich glioma-inactivated protein 3) (Leubrin). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.044744 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.046568 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.050031 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.010403 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.051793 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
COVA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.018222 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
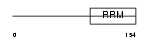
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
LSR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.041130 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KG5, Q3TJE7, Q3UIQ9, Q61148, Q61149, Q6U816 | Gene names | Lsr, Lisch7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor precursor (Liver-specific bHLH-Zip transcription factor) (Lipolysis-stimulated receptor) (Liver- specific gene on mouse chromosome 7 protein). | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.003695 (rank : 141) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
MYLK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.007981 (rank : 135) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
SFR16_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.038900 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
TROP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.008092 (rank : 134) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.055130 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.034740 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.009090 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.042555 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.029443 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.024780 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.058051 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ORC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.018107 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
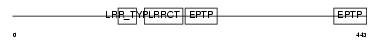
Domain Architecture |
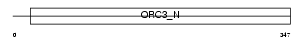
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.077074 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.021504 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.028791 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.008324 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.005861 (rank : 138) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
GAGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.024139 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75459, Q6FGM3, Q9BSS7 | Gene names | PAGE1, GAGE9, GAGEB1 | |||
|
Domain Architecture |
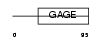
|
|||||
| Description | G antigen family B member 1 (Prostate-associated gene 1 protein) (PAGE-1) (GAGE-9) (AL5). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.048930 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
OSBL7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.011482 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.018857 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.084752 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.044296 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.018764 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.041419 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.015841 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
TMEM8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.013955 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESN3, Q99JS5 | Gene names | Tmem8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 8 precursor (M83 protein). | |||||
|
TRA2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.026106 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.026106 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.015600 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.012487 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.007311 (rank : 136) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.028697 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
LSR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.046589 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
PPIL4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.025805 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.074084 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.018954 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | -0.000297 (rank : 142) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
URIC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.037580 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25688 | Gene names | Uox | |||
|
Domain Architecture |

|
|||||
| Description | Uricase (EC 1.7.3.3) (Urate oxidase). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.022585 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
COVA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.016159 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
DLGP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.008493 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
ERCC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.020306 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
HMDH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.008966 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01237, Q5U4I2 | Gene names | Hmgcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (EC 1.1.1.34) (HMG-CoA reductase). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.036267 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.012945 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
SFR16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.062769 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.005455 (rank : 139) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SON_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.032892 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.033395 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TNR1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.005929 (rank : 137) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |
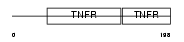
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.033368 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.061716 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.060291 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CF081_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.131951 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
CF081_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.108224 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.057705 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.055642 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.068609 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.066123 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.059150 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.073329 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.063440 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.070330 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.946010 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.934740 (rank : 3) | θ value | 3.97187e-173 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP1_MOUSE
|
||||||
| NC score | 0.902689 (rank : 4) | θ value | 2.36115e-125 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
ARMX5_HUMAN
|
||||||
| NC score | 0.833050 (rank : 5) | θ value | 2.42216e-37 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1M9, Q68DB4, Q9BVZ3, Q9H969 | Gene names | ARMCX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX5_MOUSE
|
||||||
| NC score | 0.817980 (rank : 6) | θ value | 3.61944e-33 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UZB0 | Gene names | Armcx5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.764895 (rank : 7) | θ value | 4.89182e-30 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.759135 (rank : 8) | θ value | 1.2049e-28 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX3_MOUSE
|
||||||
| NC score | 0.750388 (rank : 9) | θ value | 2.42779e-29 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX3_HUMAN
|
||||||
| NC score | 0.749073 (rank : 10) | θ value | 4.14116e-29 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.703915 (rank : 11) | θ value | 3.51386e-20 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.661993 (rank : 12) | θ value | 4.58923e-20 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ARMX6_HUMAN
|
||||||
| NC score | 0.523507 (rank : 13) | θ value | 1.43324e-05 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7L4S7, Q9NWJ3 | Gene names | ARMCX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
ARMX6_MOUSE
|
||||||
| NC score | 0.476342 (rank : 14) | θ value | 0.000786445 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3A6, Q3TUH4 | Gene names | Armcx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
ARMX4_HUMAN
|
||||||
| NC score | 0.442717 (rank : 15) | θ value | 0.000121331 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9R4, Q5H9K8, Q8N8D6 | Gene names | ARMCX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 4. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.143999 (rank : 16) | θ value | 9.29e-05 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
CF081_HUMAN
|
||||||
| NC score | 0.131951 (rank : 17) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.116713 (rank : 18) | θ value | 0.0330416 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.110653 (rank : 19) | θ value | 1.43324e-05 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CF081_MOUSE
|
||||||
| NC score | 0.108224 (rank : 20) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.104662 (rank : 21) | θ value | 0.000602161 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.103939 (rank : 22) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.101119 (rank : 23) | θ value | 4.45536e-07 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.100838 (rank : 24) | θ value | 0.0148317 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.100838 (rank : 25) | θ value | 0.0148317 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.100785 (rank : 26) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NUFP1_HUMAN
|
||||||
| NC score | 0.096994 (rank : 27) | θ value | 0.00228821 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.089482 (rank : 28) | θ value | 0.00228821 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.086225 (rank : 29) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.086055 (rank : 30) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.084752 (rank : 31) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.077074 (rank : 32) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
STRAD_MOUSE
|
||||||
| NC score | 0.077055 (rank : 33) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.074084 (rank : 34) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.073329 (rank : 35) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.072683 (rank : 36) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.071484 (rank : 37) | θ value | 0.00228821 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.071477 (rank : 38) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.071477 (rank : 39) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.070973 (rank : 40) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.070330 (rank : 41) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.068609 (rank : 42) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.066123 (rank : 43) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.063440 (rank : 44) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.062769 (rank : 45) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.061716 (rank : 46) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
YIPF2_MOUSE
|
||||||
| NC score | 0.061639 (rank : 47) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LP8, Q99LF3 | Gene names | Yipf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF2 (YIP1 family member 2). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.060291 (rank : 48) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.059755 (rank : 49) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.059150 (rank : 50) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.058051 (rank : 51) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.057705 (rank : 52) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.055642 (rank : 53) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.055130 (rank : 54) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.051793 (rank : 55) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.050031 (rank : 56) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.048930 (rank : 57) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.048874 (rank : 58) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.047993 (rank : 59) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
K1196_HUMAN
|
||||||
| NC score | 0.047461 (rank : 60) | θ value | 0.0193708 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.047261 (rank : 61) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.047140 (rank : 62) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.047114 (rank : 63) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
LSR_HUMAN
|
||||||
| NC score | 0.046589 (rank : 64) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.046568 (rank : 65) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.046074 (rank : 66) | θ value | 0.00134147 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.044744 (rank : 67) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.044296 (rank : 68) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.043219 (rank : 69) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.042555 (rank : 70) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.041419 (rank : 71) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
GNAS1_HUMAN
|
||||||
| NC score | 0.041411 (rank : 72) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
LSR_MOUSE
|
||||||
| NC score | 0.041130 (rank : 73) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KG5, Q3TJE7, Q3UIQ9, Q61148, Q61149, Q6U816 | Gene names | Lsr, Lisch7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor precursor (Liver-specific bHLH-Zip transcription factor) (Lipolysis-stimulated receptor) (Liver- specific gene on mouse chromosome 7 protein). | |||||
|
SFR16_HUMAN
|
||||||
| NC score | 0.038900 (rank : 74) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
URIC_MOUSE
|
||||||
| NC score | 0.037580 (rank : 75) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25688 | Gene names | Uox | |||
|
Domain Architecture |

|
|||||
| Description | Uricase (EC 1.7.3.3) (Urate oxidase). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.037108 (rank : 76) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.036267 (rank : 77) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.035959 (rank : 78) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.034740 (rank : 79) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.034688 (rank : 80) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.033395 (rank : 81) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.033368 (rank : 82) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
XPC_HUMAN
|
||||||
| NC score | 0.033333 (rank : 83) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01831, Q96AX0 | Gene names | XPC, XPCC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells (Xeroderma pigmentosum group C-complementing protein) (p125). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.032892 (rank : 84) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.031007 (rank : 85) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.030660 (rank : 86) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
ERCC2_HUMAN
|
||||||
| NC score | 0.030452 (rank : 87) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
LC7L2_HUMAN
|
||||||
| NC score | 0.030329 (rank : 88) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.029443 (rank : 89) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.028791 (rank : 90) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.028697 (rank : 91) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.028366 (rank : 92) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
CC054_MOUSE
|
||||||
| NC score | 0.027164 (rank : 93) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX62, Q80WB2, Q8BPE9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf54 homolog. | |||||
|
TRA2B_HUMAN
|
||||||
| NC score | 0.026106 (rank : 94) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| NC score | 0.026106 (rank : 95) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
PPIL4_MOUSE
|
||||||
| NC score | 0.025805 (rank : 96) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.025457 (rank : 97) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.024780 (rank : 98) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
GAGB1_HUMAN
|
||||||
| NC score | 0.024139 (rank : 99) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75459, Q6FGM3, Q9BSS7 | Gene names | PAGE1, GAGE9, GAGEB1 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen family B member 1 (Prostate-associated gene 1 protein) (PAGE-1) (GAGE-9) (AL5). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.023966 (rank : 100) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.022585 (rank : 101) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.021504 (rank : 102) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ERCC2_MOUSE
|
||||||
| NC score | 0.020306 (rank : 103) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.018954 (rank : 104) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.018857 (rank : 105) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.018841 (rank : 106) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.018764 (rank : 107) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PHF8_HUMAN
|
||||||
| NC score | 0.018679 (rank : 108) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.018222 (rank : 109) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
ORC3_MOUSE
|
||||||
| NC score | 0.018107 (rank : 110) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.017944 (rank : 111) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.017672 (rank : 112) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
COVA1_MOUSE
|
||||||
| NC score | 0.016159 (rank : 113) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.015841 (rank : 114) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.015600 (rank : 115) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.015368 (rank : 116) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
PAX1_HUMAN
|
||||||
| NC score | 0.014954 (rank : 117) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15863, Q642X9, Q6NTC0, Q9Y558 | Gene names | PAX1, HUP48 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-1 (HUP48). | |||||
|
TMEM8_MOUSE
|
||||||
| NC score | 0.013955 (rank : 118) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESN3, Q99JS5 | Gene names | Tmem8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 8 precursor (M83 protein). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.013863 (rank : 119) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.012945 (rank : 120) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.012487 (rank : 121) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.011813 (rank : 122) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.011595 (rank : 123) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
OSBL7_HUMAN
|
||||||
| NC score | 0.011482 (rank : 124) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
LGI3_MOUSE
|
||||||
| NC score | 0.010836 (rank : 125) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K406 | Gene names | Lgi3 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat LGI family member 3 precursor (Leucine-rich glioma-inactivated protein 3) (Leubrin). | |||||
|
LGI3_HUMAN
|
||||||
| NC score | 0.010748 (rank : 126) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N145, Q86TL4, Q8N296 | Gene names | LGI3, LGIL4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat LGI family member 3 precursor (Leucine-rich glioma-inactivated protein 3) (LGI1-like protein 4). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.010403 (rank : 127) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.009915 (rank : 128) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.009090 (rank : 129) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
HMDH_MOUSE
|
||||||
| NC score | 0.008966 (rank : 130) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01237, Q5U4I2 | Gene names | Hmgcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (EC 1.1.1.34) (HMG-CoA reductase). | |||||
|
DLGP4_HUMAN
|
||||||
| NC score | 0.008493 (rank : 131) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.008324 (rank : 132) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.008263 (rank : 133) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.008092 (rank : 134) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MYLK_HUMAN
|
||||||
| NC score | 0.007981 (rank : 135) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.007311 (rank : 136) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
TNR1B_MOUSE
|
||||||
| NC score | 0.005929 (rank : 137) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.005861 (rank : 138) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.005455 (rank : 139) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
M3K4_HUMAN
|
||||||
| NC score | 0.004164 (rank : 140) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6R4, Q92612 | Gene names | MAP3K4, KIAA0213, MAPKKK4, MEKK4, MTK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4) (MAP three kinase 1). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | 0.003695 (rank : 141) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | -0.000297 (rank : 142) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||