Please be patient as the page loads
|
THAP4_MOUSE
|
||||||
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
THAP4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.959469 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
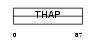
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP1_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 3) | NC score | 0.668262 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHW1 | Gene names | Thap1 | |||
|
Domain Architecture |
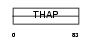
|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP1_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 4) | NC score | 0.670593 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NVV9, Q53FQ1, Q6IA99 | Gene names | THAP1 | |||
|
Domain Architecture |
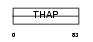
|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP8_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 5) | NC score | 0.681046 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |
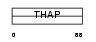
|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
THAP2_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 6) | NC score | 0.621709 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0W7 | Gene names | THAP2 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP2_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 7) | NC score | 0.623010 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP3_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 8) | NC score | 0.601335 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP3_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 9) | NC score | 0.595908 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJ25, Q8BII6 | Gene names | Thap3 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP7_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 10) | NC score | 0.506317 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 11) | NC score | 0.506119 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP6_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 12) | NC score | 0.548912 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
P52K_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 13) | NC score | 0.268544 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
P52K_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 14) | NC score | 0.274226 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.032884 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 16) | NC score | 0.025363 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.064983 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.058525 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.074516 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TATD2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.085671 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.017528 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.034128 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.037399 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.032897 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.017612 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.063827 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.057278 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYBPH_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.025188 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |
|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.065780 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
K0562_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.042459 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.033505 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.031132 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.023965 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
41_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.016250 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.058598 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.025957 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.060118 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.026119 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TEX2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.039101 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
CQ028_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.049808 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.012562 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.046494 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.038060 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
IF3X_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.040840 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
N4BP3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.055678 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15049, Q7Z6I3 | Gene names | N4BP3, KIAA0341 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.031201 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
ZBT37_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.005448 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TC79, Q5TC80, Q96M87, Q9BQ88 | Gene names | ZBTB37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 37. | |||||
|
ZSCA5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.004056 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.026494 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
DLGP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.023472 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
MEF2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.022763 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |
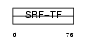
|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.008720 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.053256 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.054770 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.057909 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.054993 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.030327 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
ABCF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.020879 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
ASPX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.046710 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
CQ028_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.046086 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1F6, Q8C8V6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 homolog. | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.019784 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.034573 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.018177 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.020164 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.049002 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.021232 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.026629 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
TBX4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.009388 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |
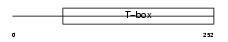
|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
TNR8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.024860 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
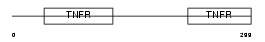
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
TOPK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.006976 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.058620 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.014597 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.027454 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.015408 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.033984 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.006511 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.020900 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.022516 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
LZTS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.038956 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.023050 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.011634 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.033797 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.053843 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.015841 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.015087 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
K1688_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.016624 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.021601 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.023602 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NP1L3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.027581 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
PHF8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.015495 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.014082 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.060467 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCAL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.018121 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.035589 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.042094 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.038805 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ZIC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.000964 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60481 | Gene names | ZIC3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein ZIC 3 (Zinc finger protein of the cerebellum 3). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.031768 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.015958 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.032179 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CACB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.013366 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |
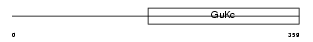
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
CACB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.021130 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |
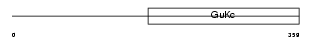
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
CLCB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.019392 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.027481 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.008202 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.009446 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.014095 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.017482 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LYAR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.020695 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.028566 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.028821 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.045353 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.050170 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
UB2E3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.007952 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969T4, Q5U0R7, Q7Z4W4 | Gene names | UBE2E3, UBCE4, UBCH9 | |||
|
Domain Architecture |
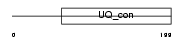
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcH9) (UbcM2). | |||||
|
UB2E3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.007952 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52483, O09180, Q3TI00, Q91X63 | Gene names | Ube2e3, Ubce4, Ubcm2 | |||
|
Domain Architecture |
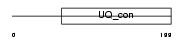
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcM2). | |||||
|
XRCC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.029552 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.005362 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.011882 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.021660 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CLCB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.018714 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.037201 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.012152 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.016232 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
NUPL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.015218 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R332, Q3UJF4, Q8BUA7, Q8BVG7, Q8C0W3 | Gene names | Nupl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.012802 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.005581 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
REST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.013808 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.021973 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.023701 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.030123 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
THA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.121943 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |
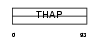
|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
THA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.076562 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK4, O94795 | Gene names | THAP11 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
THA11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.066509 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJD0 | Gene names | Thap11 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.959469 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP8_HUMAN
|
||||||
| NC score | 0.681046 (rank : 3) | θ value | 4.91457e-14 (rank : 5) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
THAP1_HUMAN
|
||||||
| NC score | 0.670593 (rank : 4) | θ value | 5.8054e-15 (rank : 4) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NVV9, Q53FQ1, Q6IA99 | Gene names | THAP1 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP1_MOUSE
|
||||||
| NC score | 0.668262 (rank : 5) | θ value | 4.44505e-15 (rank : 3) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHW1 | Gene names | Thap1 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP2_MOUSE
|
||||||
| NC score | 0.623010 (rank : 6) | θ value | 3.89403e-11 (rank : 7) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP2_HUMAN
|
||||||
| NC score | 0.621709 (rank : 7) | θ value | 2.28291e-11 (rank : 6) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0W7 | Gene names | THAP2 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP3_HUMAN
|
||||||
| NC score | 0.601335 (rank : 8) | θ value | 9.59137e-10 (rank : 8) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP3_MOUSE
|
||||||
| NC score | 0.595908 (rank : 9) | θ value | 8.11959e-09 (rank : 9) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJ25, Q8BII6 | Gene names | Thap3 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP6_HUMAN
|
||||||
| NC score | 0.548912 (rank : 10) | θ value | 1.17247e-07 (rank : 12) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
THAP7_MOUSE
|
||||||
| NC score | 0.506317 (rank : 11) | θ value | 1.80886e-08 (rank : 10) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.506119 (rank : 12) | θ value | 4.0297e-08 (rank : 11) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
P52K_MOUSE
|
||||||
| NC score | 0.274226 (rank : 13) | θ value | 0.00298849 (rank : 14) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.268544 (rank : 14) | θ value | 0.00298849 (rank : 13) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
THA10_HUMAN
|
||||||
| NC score | 0.121943 (rank : 15) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
TATD2_HUMAN
|
||||||
| NC score | 0.085671 (rank : 16) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
THA11_HUMAN
|
||||||
| NC score | 0.076562 (rank : 17) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK4, O94795 | Gene names | THAP11 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.074516 (rank : 18) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
THA11_MOUSE
|
||||||
| NC score | 0.066509 (rank : 19) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJD0 | Gene names | Thap11 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.065780 (rank : 20) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.064983 (rank : 21) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.063827 (rank : 22) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.060467 (rank : 23) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.060118 (rank : 24) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.058620 (rank : 25) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.058598 (rank : 26) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.058525 (rank : 27) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.057909 (rank : 28) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.057278 (rank : 29) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
N4BP3_HUMAN
|
||||||
| NC score | 0.055678 (rank : 30) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15049, Q7Z6I3 | Gene names | N4BP3, KIAA0341 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.054993 (rank : 31) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.054770 (rank : 32) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.053843 (rank : 33) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.053256 (rank : 34) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.050170 (rank : 35) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CQ028_HUMAN
|
||||||
| NC score | 0.049808 (rank : 36) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.049002 (rank : 37) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ASPX_MOUSE
|
||||||
| NC score | 0.046710 (rank : 38) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.046494 (rank : 39) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
CQ028_MOUSE
|
||||||
| NC score | 0.046086 (rank : 40) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1F6, Q8C8V6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 homolog. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.045353 (rank : 41) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
K0562_HUMAN
|
||||||
| NC score | 0.042459 (rank : 42) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.042094 (rank : 43) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
IF3X_HUMAN
|
||||||
| NC score | 0.040840 (rank : 44) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
TEX2_MOUSE
|
||||||
| NC score | 0.039101 (rank : 45) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
LZTS1_HUMAN
|
||||||
| NC score | 0.038956 (rank : 46) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.038805 (rank : 47) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.038060 (rank : 48) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.037399 (rank : 49) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.037201 (rank : 50) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.035589 (rank : 51) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.034573 (rank : 52) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.034128 (rank : 53) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.033984 (rank : 54) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.033797 (rank : 55) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.033505 (rank : 56) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.032897 (rank : 57) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.032884 (rank : 58) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.032179 (rank : 59) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.031768 (rank : 60) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.031201 (rank : 61) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.031132 (rank : 62) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.030327 (rank : 63) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.030123 (rank : 64) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
XRCC1_HUMAN
|
||||||
| NC score | 0.029552 (rank : 65) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.028821 (rank : 66) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.028566 (rank : 67) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NP1L3_HUMAN
|
||||||
| NC score | 0.027581 (rank : 68) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.027481 (rank : 69) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.027454 (rank : 70) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.026629 (rank : 71) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.026494 (rank : 72) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.026119 (rank : 73) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.025957 (rank : 74) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.025363 (rank : 75) | θ value | 0.0431538 (rank : 16) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MYBPH_MOUSE
|
||||||
| NC score | 0.025188 (rank : 76) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.024860 (rank : 77) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.023965 (rank : 78) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.023701 (rank : 79) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.023602 (rank : 80) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DLGP3_HUMAN
|
||||||
| NC score | 0.023472 (rank : 81) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.023050 (rank : 82) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
MEF2A_HUMAN
|
||||||
| NC score | 0.022763 (rank : 83) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.022516 (rank : 84) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.021973 (rank : 85) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.021660 (rank : 86) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.021601 (rank : 87) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.021232 (rank : 88) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CACB3_MOUSE
|
||||||
| NC score | 0.021130 (rank : 89) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.020900 (rank : 90) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
ABCF1_MOUSE
|
||||||
| NC score | 0.020879 (rank : 91) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
LYAR_MOUSE
|
||||||
| NC score | 0.020695 (rank : 92) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.020164 (rank : 93) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.019784 (rank : 94) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
CLCB_MOUSE
|
||||||
| NC score | 0.019392 (rank : 95) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
CLCB_HUMAN
|
||||||
| NC score | 0.018714 (rank : 96) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.018177 (rank : 97) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TCAL4_HUMAN
|
||||||
| NC score | 0.018121 (rank : 98) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.017612 (rank : 99) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.017528 (rank : 100) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.017482 (rank : 101) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.016624 (rank : 102) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
41_MOUSE
|
||||||
| NC score | 0.016250 (rank : 103) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.016232 (rank : 104) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.015958 (rank : 105) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.015841 (rank : 106) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
PHF8_HUMAN
|
||||||
| NC score | 0.015495 (rank : 107) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.015408 (rank : 108) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
NUPL1_MOUSE
|
||||||
| NC score | 0.015218 (rank : 109) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R332, Q3UJF4, Q8BUA7, Q8BVG7, Q8C0W3 | Gene names | Nupl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin p58/p45 (Nucleoporin-like 1). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.015087 (rank : 110) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.014597 (rank : 111) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.014095 (rank : 112) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.014082 (rank : 113) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.013808 (rank : 114) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CACB3_HUMAN
|
||||||
| NC score | 0.013366 (rank : 115) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.012802 (rank : 116) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.012562 (rank : 117) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
NGAP_HUMAN
|
||||||
| NC score | 0.012152 (rank : 118) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.011882 (rank : 119) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.011634 (rank : 120) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.009446 (rank : 121) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
TBX4_HUMAN
|
||||||
| NC score | 0.009388 (rank : 122) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.008720 (rank : 123) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.008202 (rank : 124) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
UB2E3_HUMAN
|
||||||
| NC score | 0.007952 (rank : 125) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969T4, Q5U0R7, Q7Z4W4 | Gene names | UBE2E3, UBCE4, UBCH9 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcH9) (UbcM2). | |||||
|
UB2E3_MOUSE
|
||||||
| NC score | 0.007952 (rank : 126) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52483, O09180, Q3TI00, Q91X63 | Gene names | Ube2e3, Ubce4, Ubcm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcM2). | |||||
|
TOPK_MOUSE
|
||||||
| NC score | 0.006976 (rank : 127) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.006511 (rank : 128) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.005581 (rank : 129) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
ZBT37_HUMAN
|
||||||
| NC score | 0.005448 (rank : 130) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TC79, Q5TC80, Q96M87, Q9BQ88 | Gene names | ZBTB37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 37. | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.005362 (rank : 131) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ZSCA5_HUMAN
|
||||||
| NC score | 0.004056 (rank : 132) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
ZIC3_HUMAN
|
||||||
| NC score | 0.000964 (rank : 133) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 130 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60481 | Gene names | ZIC3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein ZIC 3 (Zinc finger protein of the cerebellum 3). | |||||