Please be patient as the page loads
|
PARP4_HUMAN
|
||||||
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PARP4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | 3.85808e-43 (rank : 2) | NC score | 0.641603 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 3) | NC score | 0.630753 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
PARP1_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 4) | NC score | 0.557600 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 5) | NC score | 0.548923 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP2_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 6) | NC score | 0.566220 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PARP2_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 7) | NC score | 0.551828 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP3_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 8) | NC score | 0.538458 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 9) | NC score | 0.344964 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 10) | NC score | 0.287804 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 11) | NC score | 0.280170 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 12) | NC score | 0.076273 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 13) | NC score | 0.081017 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 14) | NC score | 0.275438 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 15) | NC score | 0.286447 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 16) | NC score | 0.059614 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 17) | NC score | 0.106440 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 18) | NC score | 0.106422 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 19) | NC score | 0.247242 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
TSH1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.038140 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
CCDC4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.102755 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
CN032_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.098698 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.227211 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.025725 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.048274 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ZCC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.062668 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
M3K6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.003583 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.031686 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.005921 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.033473 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
EGLX_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.036814 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG58, Q8CAF1, Q9D499 | Gene names | Ph4 | |||
|
Domain Architecture |
|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.022006 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.043955 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.067382 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.053838 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.033182 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.038472 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.041826 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.011741 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.023957 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.023706 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.024525 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
ANKZ1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.034562 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
CO039_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.050974 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.032884 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.012034 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.027027 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.030072 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
BACH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.025951 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00154, O43703, Q5JYL4, Q9UJM9, Q9Y539, Q9Y540 | Gene names | ACOT7, BACH | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
BT3A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.013672 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78410, O00477, O15338, O75658, Q76PA0, Q9BU81, Q9NR44 | Gene names | BTN3A2, BT3.2, BTF3, BTF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 3 member A2 precursor. | |||||
|
CO039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.052538 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.009946 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
FA8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.012746 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
GP176_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.007444 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.019549 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.023599 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.031118 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NRG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.031274 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35181 | Gene names | Nrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
SMRCD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.009913 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.008249 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
K0773_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.023082 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XD5, Q7L0D6, Q86T97 | Gene names | KIAA0773 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein KIAA0773. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.028481 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.023039 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
STAR7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.028686 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.047266 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
HDAC7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.011038 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
HSF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.019695 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.056935 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.022409 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
SL9A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.012349 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48764 | Gene names | SLC9A3, NHE3 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 3 (Na(+)/H(+) exchanger 3) (NHE-3) (Solute carrier family 9 member 3). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.022948 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.017815 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
CBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.014220 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.010452 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.019899 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.009137 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
GBP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.012019 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.007144 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
SP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.003540 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.069817 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.069384 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
FKSG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.014233 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU6 | Gene names | FKSG2 | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis inhibitor FKSG2. | |||||
|
IF35_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.022681 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.014960 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.014892 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
K0100_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.017532 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14667, Q3SYN5, Q49A07, Q5H9T4, Q6WG74, Q6ZP51, Q96HH8 | Gene names | KIAA0100, BCOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0378 family protein KIAA0100 precursor (Breast cancer overexpressed gene 1 protein) (Antigen MLAA-22). | |||||
|
K0100_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.017805 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SYL3, Q3UE65, Q66JY1, Q6DIC4, Q80U77, Q8C1W6, Q8CE06, Q8CGE3, Q9CSS8 | Gene names | Kiaa0100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0378 family protein KIAA0100 precursor. | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.020000 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RNPC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.007798 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
STAR7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.022773 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.013102 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TAF5L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.001983 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75529, Q8IW31 | Gene names | TAF5L, PAF65B | |||
|
Domain Architecture |

|
|||||
| Description | TAF5-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 5L (PCAF-associated factor 65 beta) (PAF65- beta). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.012802 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
RPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.052941 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.641603 (rank : 2) | θ value | 3.85808e-43 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.630753 (rank : 3) | θ value | 5.76516e-39 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
PARP2_HUMAN
|
||||||
| NC score | 0.566220 (rank : 4) | θ value | 3.75424e-22 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PARP1_HUMAN
|
||||||
| NC score | 0.557600 (rank : 5) | θ value | 3.07116e-24 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP2_MOUSE
|
||||||
| NC score | 0.551828 (rank : 6) | θ value | 5.07402e-19 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.548923 (rank : 7) | θ value | 4.43474e-23 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP3_HUMAN
|
||||||
| NC score | 0.538458 (rank : 8) | θ value | 1.09485e-13 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.344964 (rank : 9) | θ value | 5.08577e-11 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.287804 (rank : 10) | θ value | 2.21117e-06 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.286447 (rank : 11) | θ value | 0.000602161 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.280170 (rank : 12) | θ value | 2.44474e-05 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.275438 (rank : 13) | θ value | 0.000270298 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.247242 (rank : 14) | θ value | 0.00175202 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.227211 (rank : 15) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.106440 (rank : 16) | θ value | 0.00134147 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.106422 (rank : 17) | θ value | 0.00134147 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CCDC4_HUMAN
|
||||||
| NC score | 0.102755 (rank : 18) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.098698 (rank : 19) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.081017 (rank : 20) | θ value | 5.44631e-05 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.076273 (rank : 21) | θ value | 3.19293e-05 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.069817 (rank : 22) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.069384 (rank : 23) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.067382 (rank : 24) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
ZCC2_HUMAN
|
||||||
| NC score | 0.062668 (rank : 25) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.059614 (rank : 26) | θ value | 0.00134147 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.056935 (rank : 27) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.053838 (rank : 28) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
RPC1_HUMAN
|
||||||
| NC score | 0.052941 (rank : 29) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.052538 (rank : 30) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.050974 (rank : 31) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.048274 (rank : 32) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.047266 (rank : 33) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.043955 (rank : 34) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.041826 (rank : 35) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.038472 (rank : 36) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TSH1_HUMAN
|
||||||
| NC score | 0.038140 (rank : 37) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
EGLX_MOUSE
|
||||||
| NC score | 0.036814 (rank : 38) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG58, Q8CAF1, Q9D499 | Gene names | Ph4 | |||
|
Domain Architecture |

|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
ANKZ1_MOUSE
|
||||||
| NC score | 0.034562 (rank : 39) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.033473 (rank : 40) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.033182 (rank : 41) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.032884 (rank : 42) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.031686 (rank : 43) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NRG3_MOUSE
|
||||||
| NC score | 0.031274 (rank : 44) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35181 | Gene names | Nrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.031118 (rank : 45) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.030072 (rank : 46) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
STAR7_HUMAN
|
||||||
| NC score | 0.028686 (rank : 47) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.028481 (rank : 48) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.027027 (rank : 49) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BACH_HUMAN
|
||||||
| NC score | 0.025951 (rank : 50) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00154, O43703, Q5JYL4, Q9UJM9, Q9Y539, Q9Y540 | Gene names | ACOT7, BACH | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.025725 (rank : 51) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.024525 (rank : 52) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.023957 (rank : 53) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.023706 (rank : 54) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.023599 (rank : 55) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
K0773_HUMAN
|
||||||
| NC score | 0.023082 (rank : 56) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XD5, Q7L0D6, Q86T97 | Gene names | KIAA0773 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein KIAA0773. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.023039 (rank : 57) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.022948 (rank : 58) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
STAR7_MOUSE
|
||||||
| NC score | 0.022773 (rank : 59) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.022681 (rank : 60) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.022409 (rank : 61) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.022006 (rank : 62) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.020000 (rank : 63) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.019899 (rank : 64) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
HSF2_HUMAN
|
||||||
| NC score | 0.019695 (rank : 65) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.019549 (rank : 66) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.017815 (rank : 67) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
K0100_MOUSE
|
||||||
| NC score | 0.017805 (rank : 68) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SYL3, Q3UE65, Q66JY1, Q6DIC4, Q80U77, Q8C1W6, Q8CE06, Q8CGE3, Q9CSS8 | Gene names | Kiaa0100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0378 family protein KIAA0100 precursor. | |||||
|
K0100_HUMAN
|
||||||
| NC score | 0.017532 (rank : 69) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14667, Q3SYN5, Q49A07, Q5H9T4, Q6WG74, Q6ZP51, Q96HH8 | Gene names | KIAA0100, BCOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0378 family protein KIAA0100 precursor (Breast cancer overexpressed gene 1 protein) (Antigen MLAA-22). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.014960 (rank : 70) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.014892 (rank : 71) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
FKSG2_HUMAN
|
||||||
| NC score | 0.014233 (rank : 72) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU6 | Gene names | FKSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis inhibitor FKSG2. | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.014220 (rank : 73) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
BT3A2_HUMAN
|
||||||
| NC score | 0.013672 (rank : 74) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78410, O00477, O15338, O75658, Q76PA0, Q9BU81, Q9NR44 | Gene names | BTN3A2, BT3.2, BTF3, BTF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 3 member A2 precursor. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.013102 (rank : 75) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.012802 (rank : 76) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.012746 (rank : 77) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
SL9A3_HUMAN
|
||||||
| NC score | 0.012349 (rank : 78) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48764 | Gene names | SLC9A3, NHE3 | |||
|
Domain Architecture |
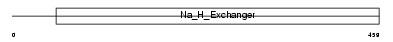
|
|||||
| Description | Sodium/hydrogen exchanger 3 (Na(+)/H(+) exchanger 3) (NHE-3) (Solute carrier family 9 member 3). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.012034 (rank : 79) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
GBP2_HUMAN
|
||||||
| NC score | 0.012019 (rank : 80) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.011741 (rank : 81) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
HDAC7_MOUSE
|
||||||
| NC score | 0.011038 (rank : 82) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.010452 (rank : 83) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.009946 (rank : 84) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
SMRCD_HUMAN
|
||||||
| NC score | 0.009913 (rank : 85) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.009137 (rank : 86) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.008249 (rank : 87) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
RNPC1_MOUSE
|
||||||
| NC score | 0.007798 (rank : 88) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
GP176_MOUSE
|
||||||
| NC score | 0.007444 (rank : 89) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.007144 (rank : 90) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.005921 (rank : 91) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
M3K6_HUMAN
|
||||||
| NC score | 0.003583 (rank : 92) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.003540 (rank : 93) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
TAF5L_HUMAN
|
||||||
| NC score | 0.001983 (rank : 94) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75529, Q8IW31 | Gene names | TAF5L, PAF65B | |||
|
Domain Architecture |

|
|||||
| Description | TAF5-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 5L (PCAF-associated factor 65 beta) (PAF65- beta). | |||||