Please be patient as the page loads
|
CAC2D_MOUSE
|
||||||
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAC2D_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997479 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 3) | NC score | 0.360114 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 4) | NC score | 0.350310 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 5) | NC score | 0.312839 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 6) | NC score | 0.307037 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 7) | NC score | 0.305760 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 8) | NC score | 0.306030 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 9) | NC score | 0.311689 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
MATN1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 10) | NC score | 0.147159 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 19 | |
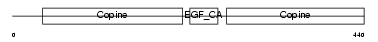
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 11) | NC score | 0.098544 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COEA1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 12) | NC score | 0.099513 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.094250 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
MATN1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.136775 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 20 | |
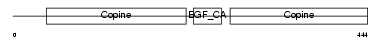
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.092268 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
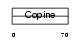
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
COEA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.087458 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.089588 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
SF04_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.065432 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
MATN2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.063225 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.083060 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 18 | |
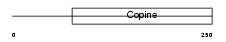
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
SASH1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.040463 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
SF04_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.046655 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
MATN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.060784 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.071679 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
NDUBB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.075081 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NX14, Q5JRR3, Q5JRR4, Q6IAB6, Q8WZ96, Q9BXX9 | Gene names | NDUFB11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase ESSS subunit) (Complex I-ESSS) (CI-ESSS) (Neuronal protein 17.3) (p17.3) (Np17.3). | |||||
|
MATN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.074808 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.015010 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
NDUBB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.062843 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O09111 | Gene names | Ndufb11, Np15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase ESSS subunit) (Complex I-ESSS) (CI-ESSS) (Neuronal protein 15.6) (p15.6) (Np15.6). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.013056 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.014551 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
ITAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.050240 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.004843 (rank : 45) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
ASCC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.023387 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8Z1, Q3TAC2 | Gene names | Ascc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 1 (ASC-1 complex subunit p50) (Trip4 complex subunit p50). | |||||
|
CANT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.022359 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVQ1, Q7Z2J7, Q8NG05, Q8NHP0, Q9BSD5 | Gene names | CANT1, SHAPY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Soluble calcium-activated nucleotidase 1 (EC 3.6.1.6) (SCAN-1) (Apyrase homolog) (Putative NF-kappa-B-activating protein 107) (Putative MAPK-activating protein PM09). | |||||
|
K1B16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.001998 (rank : 47) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04071 | Gene names | Klk1b16, Klk-16, Klk16 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b16 precursor (EC 3.4.21.54) (Gamma- renin, submandibular gland) (mGK-16). | |||||
|
MARK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | -0.000705 (rank : 48) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.010109 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.069384 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
SASH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.022273 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.003993 (rank : 46) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCFD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.016459 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVM8, O60754, O94990, Q7Z529, Q9BZI3, Q9UNL3, Q9Y6A8 | Gene names | SCFD1, KIAA0917, STXBP1L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sec1 family domain-containing protein 1 (Syntaxin-binding protein 1- like 2) (Sly1p). | |||||
|
SCFD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.016374 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRF7, Q8BRZ2, Q8K179, Q9CXR8 | Gene names | Scfd1, Stxbp1l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sec1 family domain-containing protein 1 (Syntaxin-binding protein 1- like 2). | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.053762 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
COCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.074008 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.073906 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.054753 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
VITRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.074061 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.074590 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.997479 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.360114 (rank : 3) | θ value | 2.52405e-10 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.350310 (rank : 4) | θ value | 4.76016e-09 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.312839 (rank : 5) | θ value | 5.44631e-05 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.311689 (rank : 6) | θ value | 0.000270298 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.307037 (rank : 7) | θ value | 0.000121331 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.306030 (rank : 8) | θ value | 0.00020696 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.305760 (rank : 9) | θ value | 0.000158464 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
MATN1_HUMAN
|
||||||
| NC score | 0.147159 (rank : 10) | θ value | 0.00228821 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_MOUSE
|
||||||
| NC score | 0.136775 (rank : 11) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.099513 (rank : 12) | θ value | 0.0113563 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.098544 (rank : 13) | θ value | 0.00869519 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.094250 (rank : 14) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
MATN3_HUMAN
|
||||||
| NC score | 0.092268 (rank : 15) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.089588 (rank : 16) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
COEA1_HUMAN
|
||||||
| NC score | 0.087458 (rank : 17) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
MATN3_MOUSE
|
||||||
| NC score | 0.083060 (rank : 18) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
NDUBB_HUMAN
|
||||||
| NC score | 0.075081 (rank : 19) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NX14, Q5JRR3, Q5JRR4, Q6IAB6, Q8WZ96, Q9BXX9 | Gene names | NDUFB11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase ESSS subunit) (Complex I-ESSS) (CI-ESSS) (Neuronal protein 17.3) (p17.3) (Np17.3). | |||||
|
MATN4_MOUSE
|
||||||
| NC score | 0.074808 (rank : 20) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
VITRN_MOUSE
|
||||||
| NC score | 0.074590 (rank : 21) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_HUMAN
|
||||||
| NC score | 0.074061 (rank : 22) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
COCH_HUMAN
|
||||||
| NC score | 0.074008 (rank : 23) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_MOUSE
|
||||||
| NC score | 0.073906 (rank : 24) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
MATN4_HUMAN
|
||||||
| NC score | 0.071679 (rank : 25) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.069384 (rank : 26) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.065432 (rank : 27) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.063225 (rank : 28) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
NDUBB_MOUSE
|
||||||
| NC score | 0.062843 (rank : 29) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O09111 | Gene names | Ndufb11, Np15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase ESSS subunit) (Complex I-ESSS) (CI-ESSS) (Neuronal protein 15.6) (p15.6) (Np15.6). | |||||
|
MATN2_MOUSE
|
||||||
| NC score | 0.060784 (rank : 30) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.054753 (rank : 31) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.053762 (rank : 32) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
ITAM_MOUSE
|
||||||
| NC score | 0.050240 (rank : 33) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.046655 (rank : 34) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
SASH1_MOUSE
|
||||||
| NC score | 0.040463 (rank : 35) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
ASCC1_MOUSE
|
||||||
| NC score | 0.023387 (rank : 36) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8Z1, Q3TAC2 | Gene names | Ascc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 1 (ASC-1 complex subunit p50) (Trip4 complex subunit p50). | |||||
|
CANT1_HUMAN
|
||||||
| NC score | 0.022359 (rank : 37) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVQ1, Q7Z2J7, Q8NG05, Q8NHP0, Q9BSD5 | Gene names | CANT1, SHAPY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Soluble calcium-activated nucleotidase 1 (EC 3.6.1.6) (SCAN-1) (Apyrase homolog) (Putative NF-kappa-B-activating protein 107) (Putative MAPK-activating protein PM09). | |||||
|
SASH1_HUMAN
|
||||||
| NC score | 0.022273 (rank : 38) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
SCFD1_HUMAN
|
||||||
| NC score | 0.016459 (rank : 39) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVM8, O60754, O94990, Q7Z529, Q9BZI3, Q9UNL3, Q9Y6A8 | Gene names | SCFD1, KIAA0917, STXBP1L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sec1 family domain-containing protein 1 (Syntaxin-binding protein 1- like 2) (Sly1p). | |||||
|
SCFD1_MOUSE
|
||||||
| NC score | 0.016374 (rank : 40) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRF7, Q8BRZ2, Q8K179, Q9CXR8 | Gene names | Scfd1, Stxbp1l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sec1 family domain-containing protein 1 (Syntaxin-binding protein 1- like 2). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.015010 (rank : 41) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.014551 (rank : 42) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.013056 (rank : 43) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.010109 (rank : 44) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.004843 (rank : 45) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.003993 (rank : 46) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
K1B16_MOUSE
|
||||||
| NC score | 0.001998 (rank : 47) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04071 | Gene names | Klk1b16, Klk-16, Klk16 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b16 precursor (EC 3.4.21.54) (Gamma- renin, submandibular gland) (mGK-16). | |||||
|
MARK3_HUMAN
|
||||||
| NC score | -0.000705 (rank : 48) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||