Please be patient as the page loads
|
SF04_HUMAN
|
||||||
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SF04_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.980317 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 3) | NC score | 0.534639 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 4) | NC score | 0.541572 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 5) | NC score | 0.348803 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 6) | NC score | 0.348467 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 7) | NC score | 0.335238 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 8) | NC score | 0.323835 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 9) | NC score | 0.323390 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 10) | NC score | 0.350818 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 11) | NC score | 0.349765 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 12) | NC score | 0.332499 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 13) | NC score | 0.329861 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
GPTC2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 14) | NC score | 0.309653 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 15) | NC score | 0.302668 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
TFP11_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 16) | NC score | 0.256668 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.251723 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
K0082_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 18) | NC score | 0.186244 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.167226 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
K0082_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.178355 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.100301 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.138097 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.033202 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.061218 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
BAT4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.168194 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
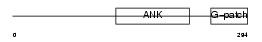
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
BAT4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.168965 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |
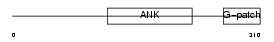
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.041807 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.028307 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.065432 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.131720 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.080084 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.231946 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
PINX1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.229804 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.011259 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.040759 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.155986 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
LRP11_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.034525 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.039118 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.038514 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.023678 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.025193 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BUB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.023447 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
MPP10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.037963 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.019609 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.026727 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
SLK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.009335 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.009396 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.140578 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.035711 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.044754 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.029677 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.026933 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
BFAR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.023929 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.019284 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CDT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.031745 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.078378 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SYCC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.019019 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.009769 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DEND_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.027673 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.019461 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.022463 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.026008 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ST2A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.013854 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50236 | Gene names | Sult2a2, Sta2, Sth2 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt sulfotransferase 2 (EC 2.8.2.14) (Hydroxysteroid sulfotransferase) (ST). | |||||
|
STRC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.016940 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.016564 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.009864 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DEND_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.030101 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
ELL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.016664 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HB65, Q9H634 | Gene names | ELL3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.091095 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.009488 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.023141 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.011558 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SPF45_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.066586 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPT6H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.034846 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
SPT6H_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.036279 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.011803 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SYIC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.016759 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.018671 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.006533 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.010977 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
NUP53_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.016071 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
OTOF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.009158 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.016960 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.109297 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
VPK10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.050729 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
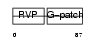
|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.050660 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050334 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050856 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050978 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.980317 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
SFR14_HUMAN
|
||||||
| NC score | 0.541572 (rank : 3) | θ value | 5.8054e-15 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.534639 (rank : 4) | θ value | 4.02038e-16 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.350818 (rank : 5) | θ value | 2.44474e-05 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.349765 (rank : 6) | θ value | 2.44474e-05 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.348803 (rank : 7) | θ value | 8.11959e-09 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.348467 (rank : 8) | θ value | 8.11959e-09 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.335238 (rank : 9) | θ value | 1.43324e-05 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.332499 (rank : 10) | θ value | 3.19293e-05 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.329861 (rank : 11) | θ value | 0.000121331 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.323835 (rank : 12) | θ value | 1.87187e-05 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.323390 (rank : 13) | θ value | 1.87187e-05 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
GPTC2_HUMAN
|
||||||
| NC score | 0.309653 (rank : 14) | θ value | 0.000786445 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| NC score | 0.302668 (rank : 15) | θ value | 0.00102713 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
TFP11_HUMAN
|
||||||
| NC score | 0.256668 (rank : 16) | θ value | 0.00228821 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.251723 (rank : 17) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.231946 (rank : 18) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
PINX1_MOUSE
|
||||||
| NC score | 0.229804 (rank : 19) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.186244 (rank : 20) | θ value | 0.0193708 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.178355 (rank : 21) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
BAT4_MOUSE
|
||||||
| NC score | 0.168965 (rank : 22) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.168194 (rank : 23) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.167226 (rank : 24) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.155986 (rank : 25) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.140578 (rank : 26) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.138097 (rank : 27) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.131720 (rank : 28) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.109297 (rank : 29) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.100301 (rank : 30) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.091095 (rank : 31) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.080084 (rank : 32) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.078378 (rank : 33) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.066586 (rank : 34) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.065432 (rank : 35) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.061218 (rank : 36) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
VPK9_HUMAN
|
||||||
| NC score | 0.050978 (rank : 37) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| NC score | 0.050856 (rank : 38) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK10_HUMAN
|
||||||
| NC score | 0.050729 (rank : 39) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| NC score | 0.050660 (rank : 40) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| NC score | 0.050334 (rank : 41) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.044754 (rank : 42) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.041807 (rank : 43) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.040759 (rank : 44) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.039118 (rank : 45) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.038514 (rank : 46) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
MPP10_HUMAN
|
||||||
| NC score | 0.037963 (rank : 47) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
SPT6H_MOUSE
|
||||||
| NC score | 0.036279 (rank : 48) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.035711 (rank : 49) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
SPT6H_HUMAN
|
||||||
| NC score | 0.034846 (rank : 50) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
LRP11_HUMAN
|
||||||
| NC score | 0.034525 (rank : 51) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.033202 (rank : 52) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CDT1_MOUSE
|
||||||
| NC score | 0.031745 (rank : 53) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
DEND_HUMAN
|
||||||
| NC score | 0.030101 (rank : 54) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.029677 (rank : 55) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.028307 (rank : 56) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
DEND_MOUSE
|
||||||
| NC score | 0.027673 (rank : 57) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.026933 (rank : 58) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.026727 (rank : 59) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.026008 (rank : 60) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.025193 (rank : 61) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BFAR_MOUSE
|
||||||
| NC score | 0.023929 (rank : 62) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.023678 (rank : 63) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BUB1_HUMAN
|
||||||
| NC score | 0.023447 (rank : 64) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.023141 (rank : 65) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.022463 (rank : 66) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.019609 (rank : 67) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.019461 (rank : 68) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.019284 (rank : 69) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
SYCC_MOUSE
|
||||||
| NC score | 0.019019 (rank : 70) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.018671 (rank : 71) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.016960 (rank : 72) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
STRC_HUMAN
|
||||||
| NC score | 0.016940 (rank : 73) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
SYIC_MOUSE
|
||||||
| NC score | 0.016759 (rank : 74) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
ELL3_HUMAN
|
||||||
| NC score | 0.016664 (rank : 75) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HB65, Q9H634 | Gene names | ELL3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.016564 (rank : 76) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
NUP53_HUMAN
|
||||||
| NC score | 0.016071 (rank : 77) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
ST2A2_MOUSE
|
||||||
| NC score | 0.013854 (rank : 78) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50236 | Gene names | Sult2a2, Sta2, Sth2 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt sulfotransferase 2 (EC 2.8.2.14) (Hydroxysteroid sulfotransferase) (ST). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.011803 (rank : 79) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.011558 (rank : 80) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.011259 (rank : 81) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.010977 (rank : 82) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.009864 (rank : 83) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.009769 (rank : 84) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.009488 (rank : 85) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.009396 (rank : 86) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.009335 (rank : 87) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
OTOF_MOUSE
|
||||||
| NC score | 0.009158 (rank : 88) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.006533 (rank : 89) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||