Please be patient as the page loads
|
K0082_HUMAN
|
||||||
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0082_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993921 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
VPK7_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 3) | NC score | 0.337166 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
RRMJ1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 4) | NC score | 0.233527 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UET6, O75670 | Gene names | FTSJ1 | |||
|
Domain Architecture |
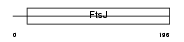
|
|||||
| Description | Putative ribosomal RNA methyltransferase 1 (EC 2.1.1.-) (rRNA (uridine-2'-O-)-methyltransferase). | |||||
|
VPK10_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.329275 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
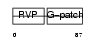
|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 6) | NC score | 0.329227 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 7) | NC score | 0.329255 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 8) | NC score | 0.329686 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 9) | NC score | 0.329545 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK9_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 10) | NC score | 0.330242 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 11) | NC score | 0.326479 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.325355 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK11_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 13) | NC score | 0.320578 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
SPF45_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.228082 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 15) | NC score | 0.228217 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
TFP11_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 16) | NC score | 0.268607 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.260606 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
VPK4_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.308307 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 19) | NC score | 0.308418 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
SF04_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.178355 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.234846 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.172262 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
STAT4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.052766 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
PINX1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.213778 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.078644 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.115569 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.135587 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
PO210_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.031484 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY81, Q3U031, Q69ZW2, Q7TQM1 | Gene names | Nup210, Kiaa0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.126951 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.157147 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.156553 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
RRMJ3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.096296 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IY81, Q8N3A3, Q8WXX1, Q9BWM4, Q9NXT6 | Gene names | FTSJ3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
STAT4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.043849 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
GPTC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.144809 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.097458 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.088231 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.134847 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.091282 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
OSBP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.016438 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.130925 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.130634 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.106530 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
FA8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.020479 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.024088 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
SFR14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.110315 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SC6A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.020785 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30531 | Gene names | SLC6A1, GABATR, GABT1, GAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent GABA transporter 1. | |||||
|
SC6A1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.020796 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31648 | Gene names | Slc6a1, Gabt1, Gat-1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent GABA transporter 1. | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.038363 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
FA8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.018979 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.132409 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
S6A12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.016488 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48065 | Gene names | SLC6A12 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent betaine transporter (Na+/Cl- betaine/GABA transporter) (BGT-1). | |||||
|
S6A12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.016583 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31651 | Gene names | Slc6a12, Gabt2, Gat-2, Gat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent betaine transporter (Na+/Cl- betaine/GABA transporter) (Sodium- and chloride-dependent GABA transporter 2) (GAT2). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.140388 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.003035 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
AT10D_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.009415 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.014662 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.015155 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
GPTC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.129811 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
INADL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.013921 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.007286 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BAT4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.065670 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
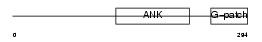
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
BAT4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056333 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |
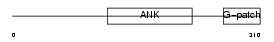
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.993921 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
VPK7_HUMAN
|
||||||
| NC score | 0.337166 (rank : 3) | θ value | 0.000158464 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK9_HUMAN
|
||||||
| NC score | 0.330242 (rank : 4) | θ value | 0.000461057 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| NC score | 0.329686 (rank : 5) | θ value | 0.000461057 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| NC score | 0.329545 (rank : 6) | θ value | 0.000461057 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK10_HUMAN
|
||||||
| NC score | 0.329275 (rank : 7) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| NC score | 0.329255 (rank : 8) | θ value | 0.000461057 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| NC score | 0.329227 (rank : 9) | θ value | 0.000461057 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| NC score | 0.326479 (rank : 10) | θ value | 0.000786445 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| NC score | 0.325355 (rank : 11) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK11_HUMAN
|
||||||
| NC score | 0.320578 (rank : 12) | θ value | 0.00175202 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| NC score | 0.308418 (rank : 13) | θ value | 0.00869519 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK4_HUMAN
|
||||||
| NC score | 0.308307 (rank : 14) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
TFP11_HUMAN
|
||||||
| NC score | 0.268607 (rank : 15) | θ value | 0.00509761 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.260606 (rank : 16) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.234846 (rank : 17) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
RRMJ1_HUMAN
|
||||||
| NC score | 0.233527 (rank : 18) | θ value | 0.00020696 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UET6, O75670 | Gene names | FTSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ribosomal RNA methyltransferase 1 (EC 2.1.1.-) (rRNA (uridine-2'-O-)-methyltransferase). | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.228217 (rank : 19) | θ value | 0.00509761 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.228082 (rank : 20) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
PINX1_MOUSE
|
||||||
| NC score | 0.213778 (rank : 21) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.178355 (rank : 22) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.172262 (rank : 23) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.157147 (rank : 24) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.156553 (rank : 25) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
GPTC2_HUMAN
|
||||||
| NC score | 0.144809 (rank : 26) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.140388 (rank : 27) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.135587 (rank : 28) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.134847 (rank : 29) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.132409 (rank : 30) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.130925 (rank : 31) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.130634 (rank : 32) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
GPTC2_MOUSE
|
||||||
| NC score | 0.129811 (rank : 33) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.126951 (rank : 34) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.115569 (rank : 35) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SFR14_HUMAN
|
||||||
| NC score | 0.110315 (rank : 36) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.106530 (rank : 37) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.097458 (rank : 38) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
RRMJ3_HUMAN
|
||||||
| NC score | 0.096296 (rank : 39) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IY81, Q8N3A3, Q8WXX1, Q9BWM4, Q9NXT6 | Gene names | FTSJ3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.091282 (rank : 40) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.088231 (rank : 41) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.078644 (rank : 42) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.065670 (rank : 43) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
BAT4_MOUSE
|
||||||
| NC score | 0.056333 (rank : 44) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
STAT4_HUMAN
|
||||||
| NC score | 0.052766 (rank : 45) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT4_MOUSE
|
||||||
| NC score | 0.043849 (rank : 46) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.038363 (rank : 47) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
PO210_MOUSE
|
||||||
| NC score | 0.031484 (rank : 48) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY81, Q3U031, Q69ZW2, Q7TQM1 | Gene names | Nup210, Kiaa0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.024088 (rank : 49) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
SC6A1_MOUSE
|
||||||
| NC score | 0.020796 (rank : 50) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31648 | Gene names | Slc6a1, Gabt1, Gat-1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent GABA transporter 1. | |||||
|
SC6A1_HUMAN
|
||||||
| NC score | 0.020785 (rank : 51) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30531 | Gene names | SLC6A1, GABATR, GABT1, GAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent GABA transporter 1. | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.020479 (rank : 52) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.018979 (rank : 53) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
S6A12_MOUSE
|
||||||
| NC score | 0.016583 (rank : 54) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31651 | Gene names | Slc6a12, Gabt2, Gat-2, Gat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent betaine transporter (Na+/Cl- betaine/GABA transporter) (Sodium- and chloride-dependent GABA transporter 2) (GAT2). | |||||
|
S6A12_HUMAN
|
||||||
| NC score | 0.016488 (rank : 55) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48065 | Gene names | SLC6A12 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent betaine transporter (Na+/Cl- betaine/GABA transporter) (BGT-1). | |||||
|
OSBP2_MOUSE
|
||||||
| NC score | 0.016438 (rank : 56) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.015155 (rank : 57) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.014662 (rank : 58) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.013921 (rank : 59) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
AT10D_MOUSE
|
||||||
| NC score | 0.009415 (rank : 60) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.007286 (rank : 61) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.003035 (rank : 62) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||