Please be patient as the page loads
|
AT10D_MOUSE
|
||||||
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AT10A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991498 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT10A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991088 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT10B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.992781 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT10D_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998484 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT8B4_HUMAN
|
||||||
| θ value | 2.52853e-119 (rank : 6) | NC score | 0.947839 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT8B2_HUMAN
|
||||||
| θ value | 1.06234e-117 (rank : 7) | NC score | 0.946238 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT8A1_MOUSE
|
||||||
| θ value | 1.8752e-114 (rank : 8) | NC score | 0.942765 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_HUMAN
|
||||||
| θ value | 5.45601e-114 (rank : 9) | NC score | 0.942946 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A2_HUMAN
|
||||||
| θ value | 2.1455e-110 (rank : 10) | NC score | 0.942488 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8A2_MOUSE
|
||||||
| θ value | 4.77967e-110 (rank : 11) | NC score | 0.942253 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT8B1_HUMAN
|
||||||
| θ value | 7.14227e-106 (rank : 12) | NC score | 0.943303 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT11C_HUMAN
|
||||||
| θ value | 2.01279e-100 (rank : 13) | NC score | 0.937140 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT11A_HUMAN
|
||||||
| θ value | 1.30465e-99 (rank : 14) | NC score | 0.934584 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11B_HUMAN
|
||||||
| θ value | 1.10445e-98 (rank : 15) | NC score | 0.940041 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT11A_MOUSE
|
||||||
| θ value | 2.72037e-97 (rank : 16) | NC score | 0.933289 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT8B3_HUMAN
|
||||||
| θ value | 2.54619e-95 (rank : 17) | NC score | 0.940929 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
ATP9B_MOUSE
|
||||||
| θ value | 2.03155e-68 (rank : 18) | NC score | 0.907403 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_HUMAN
|
||||||
| θ value | 2.65329e-68 (rank : 19) | NC score | 0.905729 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9A_HUMAN
|
||||||
| θ value | 1.11475e-66 (rank : 20) | NC score | 0.904962 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
ATP9A_MOUSE
|
||||||
| θ value | 1.04338e-64 (rank : 21) | NC score | 0.902013 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
AT2B4_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 22) | NC score | 0.558325 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT132_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 23) | NC score | 0.577225 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT133_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 24) | NC score | 0.600454 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT132_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 25) | NC score | 0.577613 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT2B2_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 26) | NC score | 0.532807 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 27) | NC score | 0.535785 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 28) | NC score | 0.537253 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 29) | NC score | 0.509684 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT12A_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 30) | NC score | 0.406582 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT12A_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 31) | NC score | 0.409979 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2C1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 32) | NC score | 0.409296 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C1_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 33) | NC score | 0.406642 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2A3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 34) | NC score | 0.405578 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A2_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 35) | NC score | 0.411054 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 36) | NC score | 0.397500 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 37) | NC score | 0.412226 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT1A2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 38) | NC score | 0.379222 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A4_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 39) | NC score | 0.386515 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT1A1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 40) | NC score | 0.380715 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 41) | NC score | 0.381364 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A2_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 42) | NC score | 0.379624 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT2A3_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 43) | NC score | 0.395427 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT1A3_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 44) | NC score | 0.361103 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 45) | NC score | 0.358589 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2A1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 46) | NC score | 0.394536 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT1A4_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 47) | NC score | 0.385669 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 48) | NC score | 0.349726 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
ATP4A_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 49) | NC score | 0.384949 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
ATP4A_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 50) | NC score | 0.389426 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT131_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 51) | NC score | 0.456515 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT131_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 52) | NC score | 0.457291 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
CYB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.040761 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00156, Q34786, Q8HBR6, Q8HNQ0, Q8HNQ1, Q8HNQ9, Q8HNR4, Q8HNR7, Q8W7V8, Q8WCV9, Q8WCY2, Q8WCY7, Q8WCY8, Q9B1A6, Q9B1B6, Q9B1B8, Q9B1D4, Q9B1X6, Q9B2V0, Q9B2V8, Q9B2W0, Q9B2W3, Q9B2W8, Q9B2X1, Q9B2X7, Q9B2X9, Q9B2Y3, Q9B2Z0, Q9B2Z4, Q9T6H6, Q9T9Y0, Q9TEH4 | Gene names | MT-CYB, COB, CYTB, MTCYB | |||
|
Domain Architecture |
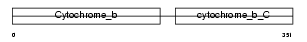
|
|||||
| Description | Cytochrome b. | |||||
|
GP128_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.013987 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96K78, Q86SQ2 | Gene names | GPR128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
CYB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.026826 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00158 | Gene names | Mt-Cyb, Cob, Cytb, mt-Cytb, Mtcyb | |||
|
Domain Architecture |
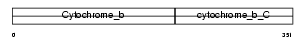
|
|||||
| Description | Cytochrome b. | |||||
|
IRK4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.005439 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52189 | Gene names | Kcnj4, Irk3 | |||
|
Domain Architecture |
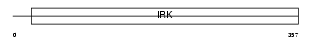
|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (IRK3). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.000705 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.004399 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
OR1N1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | -0.003724 (rank : 82) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGS0, O43870, Q6IF16, Q96R93 | Gene names | OR1N1, OR1N3 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1N1 (Olfactory receptor 1-26) (OR1-26) (Olfactory receptor 1N3) (Olfactory receptor OR9-22). | |||||
|
CS029_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.008554 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUQ7, O75229, Q7LE08, Q9BTA6, Q9Y5A4 | Gene names | C19orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 (NY-REN-24 antigen). | |||||
|
MTR1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | -0.001044 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIQ6, Q80T38, Q8K3K0 | Gene names | Mtnr1b | |||
|
Domain Architecture |
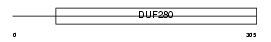
|
|||||
| Description | Melatonin receptor type 1B (Mel-1B-R) (Mel1b melatonin receptor). | |||||
|
PF2R_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.004625 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43117 | Gene names | Ptgfr | |||
|
Domain Architecture |
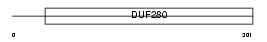
|
|||||
| Description | Prostaglandin F2-alpha receptor (Prostanoid FP receptor) (PGF receptor) (PGF2 alpha receptor). | |||||
|
C5AR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | -0.002205 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21730 | Gene names | C5AR1, C5AR, C5R1 | |||
|
Domain Architecture |
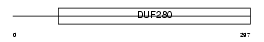
|
|||||
| Description | C5a anaphylatoxin chemotactic receptor (C5a-R) (C5aR) (CD88 antigen). | |||||
|
MBOA5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.009005 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1A2, Q7KZS1, Q92980, Q9BW40 | Gene names | MBOAT5, OACT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane bound O-acyltransferase domain-containing protein 5 (EC 2.3.-.-) (O-acyltransferase domain-containing protein 5). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.004136 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
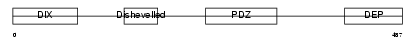
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.004139 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
IRK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.003745 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48050 | Gene names | KCNJ4 | |||
|
Domain Architecture |
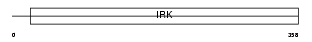
|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (Hippocampal inward rectifier) (HIR) (HRK1) (HIRK2). | |||||
|
K0082_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.009415 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.009440 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MPP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.002103 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
MRGA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.000542 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZC6 | Gene names | Mrgpra6, Mrga6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mas-related G-protein coupled receptor member A6. | |||||
|
PTX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.005815 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |
|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
SP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | -0.003531 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
ABI3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.004631 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
ARMC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.006080 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.003140 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MFN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.003908 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q811U4, Q811D5, Q8CEY6, Q99M10, Q9D395 | Gene names | Mfn1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1). | |||||
|
OR2L5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | -0.004215 (rank : 83) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG80, Q6IF04 | Gene names | OR2L5, OR2L11 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 2L5 (Olfactory receptor OR1-53). | |||||
|
TERT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.005569 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70372, O35432, Q9JK99 | Gene names | Tert | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomerase reverse transcriptase (EC 2.7.7.49) (Telomerase catalytic subunit). | |||||
|
ATP7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.147660 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.144598 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
ATP7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.151374 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.150945 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
AT10D_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_HUMAN
|
||||||
| NC score | 0.998484 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10B_HUMAN
|
||||||
| NC score | 0.992781 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT10A_HUMAN
|
||||||
| NC score | 0.991498 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT10A_MOUSE
|
||||||
| NC score | 0.991088 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT8B4_HUMAN
|
||||||
| NC score | 0.947839 (rank : 6) | θ value | 2.52853e-119 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT8B2_HUMAN
|
||||||
| NC score | 0.946238 (rank : 7) | θ value | 1.06234e-117 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT8B1_HUMAN
|
||||||
| NC score | 0.943303 (rank : 8) | θ value | 7.14227e-106 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT8A1_HUMAN
|
||||||
| NC score | 0.942946 (rank : 9) | θ value | 5.45601e-114 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_MOUSE
|
||||||
| NC score | 0.942765 (rank : 10) | θ value | 1.8752e-114 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A2_HUMAN
|
||||||
| NC score | 0.942488 (rank : 11) | θ value | 2.1455e-110 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8A2_MOUSE
|
||||||
| NC score | 0.942253 (rank : 12) | θ value | 4.77967e-110 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT8B3_HUMAN
|
||||||
| NC score | 0.940929 (rank : 13) | θ value | 2.54619e-95 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
AT11B_HUMAN
|
||||||
| NC score | 0.940041 (rank : 14) | θ value | 1.10445e-98 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT11C_HUMAN
|
||||||
| NC score | 0.937140 (rank : 15) | θ value | 2.01279e-100 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT11A_HUMAN
|
||||||
| NC score | 0.934584 (rank : 16) | θ value | 1.30465e-99 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11A_MOUSE
|
||||||
| NC score | 0.933289 (rank : 17) | θ value | 2.72037e-97 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
ATP9B_MOUSE
|
||||||
| NC score | 0.907403 (rank : 18) | θ value | 2.03155e-68 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_HUMAN
|
||||||
| NC score | 0.905729 (rank : 19) | θ value | 2.65329e-68 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9A_HUMAN
|
||||||
| NC score | 0.904962 (rank : 20) | θ value | 1.11475e-66 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
ATP9A_MOUSE
|
||||||
| NC score | 0.902013 (rank : 21) | θ value | 1.04338e-64 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
AT133_HUMAN
|
||||||
| NC score | 0.600454 (rank : 22) | θ value | 1.16975e-15 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT132_MOUSE
|
||||||
| NC score | 0.577613 (rank : 23) | θ value | 1.99529e-15 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT132_HUMAN
|
||||||
| NC score | 0.577225 (rank : 24) | θ value | 5.25075e-16 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT2B4_HUMAN
|
||||||
| NC score | 0.558325 (rank : 25) | θ value | 1.63225e-17 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.537253 (rank : 26) | θ value | 1.42992e-13 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.535785 (rank : 27) | θ value | 4.91457e-14 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| NC score | 0.532807 (rank : 28) | θ value | 3.76295e-14 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.509684 (rank : 29) | θ value | 2.28291e-11 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT131_MOUSE
|
||||||
| NC score | 0.457291 (rank : 30) | θ value | 0.0330416 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT131_HUMAN
|
||||||
| NC score | 0.456515 (rank : 31) | θ value | 0.00509761 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT2A2_MOUSE
|
||||||
| NC score | 0.412226 (rank : 32) | θ value | 5.81887e-07 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_HUMAN
|
||||||
| NC score | 0.411054 (rank : 33) | θ value | 2.61198e-07 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT12A_HUMAN
|
||||||
| NC score | 0.409979 (rank : 34) | θ value | 2.13673e-09 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2C1_HUMAN
|
||||||
| NC score | 0.409296 (rank : 35) | θ value | 2.13673e-09 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C1_MOUSE
|
||||||
| NC score | 0.406642 (rank : 36) | θ value | 8.11959e-09 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT12A_MOUSE
|
||||||
| NC score | 0.406582 (rank : 37) | θ value | 1.25267e-09 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2A3_HUMAN
|
||||||
| NC score | 0.405578 (rank : 38) | θ value | 1.99992e-07 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A1_MOUSE
|
||||||
| NC score | 0.397500 (rank : 39) | θ value | 4.45536e-07 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A3_MOUSE
|
||||||
| NC score | 0.395427 (rank : 40) | θ value | 3.19293e-05 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A1_HUMAN
|
||||||
| NC score | 0.394536 (rank : 41) | θ value | 0.00035302 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
ATP4A_MOUSE
|
||||||
| NC score | 0.389426 (rank : 42) | θ value | 0.00228821 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT1A4_HUMAN
|
||||||
| NC score | 0.386515 (rank : 43) | θ value | 8.40245e-06 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT1A4_MOUSE
|
||||||
| NC score | 0.385669 (rank : 44) | θ value | 0.000461057 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
ATP4A_HUMAN
|
||||||
| NC score | 0.384949 (rank : 45) | θ value | 0.00134147 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT1A1_MOUSE
|
||||||
| NC score | 0.381364 (rank : 46) | θ value | 1.43324e-05 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A1_HUMAN
|
||||||
| NC score | 0.380715 (rank : 47) | θ value | 1.09739e-05 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A2_HUMAN
|
||||||
| NC score | 0.379624 (rank : 48) | θ value | 1.43324e-05 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| NC score | 0.379222 (rank : 49) | θ value | 6.43352e-06 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A3_HUMAN
|
||||||
| NC score | 0.361103 (rank : 50) | θ value | 0.000158464 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| NC score | 0.358589 (rank : 51) | θ value | 0.00035302 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.349726 (rank : 52) | θ value | 0.000786445 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
ATP7B_HUMAN
|
||||||
| NC score | 0.151374 (rank : 53) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7B_MOUSE
|
||||||
| NC score | 0.150945 (rank : 54) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
ATP7A_HUMAN
|
||||||
| NC score | 0.147660 (rank : 55) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| NC score | 0.144598 (rank : 56) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
CYB_HUMAN
|
||||||
| NC score | 0.040761 (rank : 57) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00156, Q34786, Q8HBR6, Q8HNQ0, Q8HNQ1, Q8HNQ9, Q8HNR4, Q8HNR7, Q8W7V8, Q8WCV9, Q8WCY2, Q8WCY7, Q8WCY8, Q9B1A6, Q9B1B6, Q9B1B8, Q9B1D4, Q9B1X6, Q9B2V0, Q9B2V8, Q9B2W0, Q9B2W3, Q9B2W8, Q9B2X1, Q9B2X7, Q9B2X9, Q9B2Y3, Q9B2Z0, Q9B2Z4, Q9T6H6, Q9T9Y0, Q9TEH4 | Gene names | MT-CYB, COB, CYTB, MTCYB | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b. | |||||
|
CYB_MOUSE
|
||||||
| NC score | 0.026826 (rank : 58) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00158 | Gene names | Mt-Cyb, Cob, Cytb, mt-Cytb, Mtcyb | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b. | |||||
|
GP128_HUMAN
|
||||||
| NC score | 0.013987 (rank : 59) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96K78, Q86SQ2 | Gene names | GPR128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.009440 (rank : 60) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.009415 (rank : 61) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MBOA5_HUMAN
|
||||||
| NC score | 0.009005 (rank : 62) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1A2, Q7KZS1, Q92980, Q9BW40 | Gene names | MBOAT5, OACT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane bound O-acyltransferase domain-containing protein 5 (EC 2.3.-.-) (O-acyltransferase domain-containing protein 5). | |||||
|
CS029_HUMAN
|
||||||
| NC score | 0.008554 (rank : 63) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUQ7, O75229, Q7LE08, Q9BTA6, Q9Y5A4 | Gene names | C19orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 (NY-REN-24 antigen). | |||||
|
ARMC6_HUMAN
|
||||||
| NC score | 0.006080 (rank : 64) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
PTX3_MOUSE
|
||||||
| NC score | 0.005815 (rank : 65) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |

|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
TERT_MOUSE
|
||||||
| NC score | 0.005569 (rank : 66) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70372, O35432, Q9JK99 | Gene names | Tert | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomerase reverse transcriptase (EC 2.7.7.49) (Telomerase catalytic subunit). | |||||
|
IRK4_MOUSE
|
||||||
| NC score | 0.005439 (rank : 67) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52189 | Gene names | Kcnj4, Irk3 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (IRK3). | |||||
|
ABI3_MOUSE
|
||||||
| NC score | 0.004631 (rank : 68) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
PF2R_MOUSE
|
||||||
| NC score | 0.004625 (rank : 69) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43117 | Gene names | Ptgfr | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin F2-alpha receptor (Prostanoid FP receptor) (PGF receptor) (PGF2 alpha receptor). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.004399 (rank : 70) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.004139 (rank : 71) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.004136 (rank : 72) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
MFN1_MOUSE
|
||||||
| NC score | 0.003908 (rank : 73) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q811U4, Q811D5, Q8CEY6, Q99M10, Q9D395 | Gene names | Mfn1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1). | |||||
|
IRK4_HUMAN
|
||||||
| NC score | 0.003745 (rank : 74) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48050 | Gene names | KCNJ4 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (Hippocampal inward rectifier) (HIR) (HRK1) (HIRK2). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.003140 (rank : 75) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MPP4_MOUSE
|
||||||
| NC score | 0.002103 (rank : 76) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.000705 (rank : 77) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
MRGA6_MOUSE
|
||||||
| NC score | 0.000542 (rank : 78) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZC6 | Gene names | Mrgpra6, Mrga6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mas-related G-protein coupled receptor member A6. | |||||
|
MTR1B_MOUSE
|
||||||
| NC score | -0.001044 (rank : 79) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIQ6, Q80T38, Q8K3K0 | Gene names | Mtnr1b | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin receptor type 1B (Mel-1B-R) (Mel1b melatonin receptor). | |||||
|
C5AR_HUMAN
|
||||||
| NC score | -0.002205 (rank : 80) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21730 | Gene names | C5AR1, C5AR, C5R1 | |||
|
Domain Architecture |

|
|||||
| Description | C5a anaphylatoxin chemotactic receptor (C5a-R) (C5aR) (CD88 antigen). | |||||
|
SP4_MOUSE
|
||||||
| NC score | -0.003531 (rank : 81) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
OR1N1_HUMAN
|
||||||
| NC score | -0.003724 (rank : 82) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGS0, O43870, Q6IF16, Q96R93 | Gene names | OR1N1, OR1N3 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1N1 (Olfactory receptor 1-26) (OR1-26) (Olfactory receptor 1N3) (Olfactory receptor OR9-22). | |||||
|
OR2L5_HUMAN
|
||||||
| NC score | -0.004215 (rank : 83) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG80, Q6IF04 | Gene names | OR2L5, OR2L11 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 2L5 (Olfactory receptor OR1-53). | |||||