Please be patient as the page loads
|
AT10B_HUMAN
|
||||||
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AT10A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991708 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT10A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992165 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT10B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT10D_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993605 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.992781 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT8B2_HUMAN
|
||||||
| θ value | 1.81207e-117 (rank : 6) | NC score | 0.950843 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT8B4_HUMAN
|
||||||
| θ value | 2.36663e-117 (rank : 7) | NC score | 0.950884 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT8A2_MOUSE
|
||||||
| θ value | 3.77839e-115 (rank : 8) | NC score | 0.945944 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT8A2_HUMAN
|
||||||
| θ value | 2.44908e-114 (rank : 9) | NC score | 0.945916 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8B1_HUMAN
|
||||||
| θ value | 1.69998e-107 (rank : 10) | NC score | 0.947104 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT8A1_HUMAN
|
||||||
| θ value | 3.78717e-107 (rank : 11) | NC score | 0.945433 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_MOUSE
|
||||||
| θ value | 4.94618e-107 (rank : 12) | NC score | 0.944993 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT11B_HUMAN
|
||||||
| θ value | 9.98939e-100 (rank : 13) | NC score | 0.941849 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT11A_HUMAN
|
||||||
| θ value | 2.30293e-96 (rank : 14) | NC score | 0.939810 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11A_MOUSE
|
||||||
| θ value | 4.49445e-92 (rank : 15) | NC score | 0.937357 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11C_HUMAN
|
||||||
| θ value | 3.80478e-91 (rank : 16) | NC score | 0.940397 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT8B3_HUMAN
|
||||||
| θ value | 4.96919e-91 (rank : 17) | NC score | 0.943576 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
ATP9B_HUMAN
|
||||||
| θ value | 2.73936e-73 (rank : 18) | NC score | 0.909303 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_MOUSE
|
||||||
| θ value | 3.57772e-73 (rank : 19) | NC score | 0.911951 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9A_HUMAN
|
||||||
| θ value | 1.00825e-67 (rank : 20) | NC score | 0.908914 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
ATP9A_MOUSE
|
||||||
| θ value | 1.90147e-66 (rank : 21) | NC score | 0.905638 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
AT2B4_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 22) | NC score | 0.551641 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 23) | NC score | 0.502749 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 24) | NC score | 0.530326 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT132_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 25) | NC score | 0.566926 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 26) | NC score | 0.529355 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 27) | NC score | 0.525539 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT132_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 28) | NC score | 0.567380 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT133_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 29) | NC score | 0.585031 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT12A_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 30) | NC score | 0.398982 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT12A_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 31) | NC score | 0.395396 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT1A4_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 32) | NC score | 0.376495 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT1A4_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 33) | NC score | 0.375949 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT2C1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 34) | NC score | 0.387377 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
ATP4A_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 35) | NC score | 0.375984 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
ATP4A_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 36) | NC score | 0.380352 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT2C1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 37) | NC score | 0.390128 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT131_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 38) | NC score | 0.455553 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT1A2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 39) | NC score | 0.369385 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 40) | NC score | 0.368993 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 41) | NC score | 0.370363 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT131_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 42) | NC score | 0.457013 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT1A1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 43) | NC score | 0.370991 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A3_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 44) | NC score | 0.350867 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 45) | NC score | 0.348456 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2A2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 46) | NC score | 0.387414 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 47) | NC score | 0.388634 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 48) | NC score | 0.330920 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
AT2A1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 49) | NC score | 0.374535 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 50) | NC score | 0.372092 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 51) | NC score | 0.381962 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 52) | NC score | 0.372414 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
PUM1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.023577 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14671, Q5VXY7, Q9HAN1 | Gene names | PUM1, KIAA0099, PUMH1 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1 (Pumilio-1) (HsPUM). | |||||
|
PUM1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.023605 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U78, Q80X96, Q80YU8, Q8BPV7, Q9EPU6 | Gene names | Pum1, Kiaa0099 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1. | |||||
|
S6A18_MOUSE
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.005037 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88576, O88577, O88578, O88579, O88580, O88581, Q91XG6 | Gene names | Slc6a18, Xt2, Xtrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent transporter XTRP2 (Solute carrier family 6 member 18). | |||||
|
TSH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.011172 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
KI20A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.006855 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95235 | Gene names | KIF20A, RAB6KIFL | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin family member 20A (Rabkinesin-6) (Rab6-interacting kinesin- like protein) (GG10_2). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | -0.000065 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RNF12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.010205 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.000752 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.009675 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.005621 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.005747 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MMEL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.011499 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLI3, Q3U495, Q9ERK2, Q9ERK3, Q9QZV6, Q9QZV7 | Gene names | Mmel1, Nep2, Nl1, Sep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) (Neprilysin-like peptidase) (NEPLP) (Neprilysin-like 1) (NL-1) (Soluble secreted endopeptidase) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.002674 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RD23B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.008805 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |
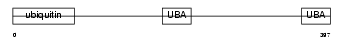
|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
KIFC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.003811 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08672, O08613 | Gene names | Kifc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC2. | |||||
|
REL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.005797 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | -0.000299 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MTR1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | -0.003236 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49286 | Gene names | MTNR1B | |||
|
Domain Architecture |
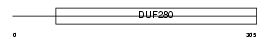
|
|||||
| Description | Melatonin receptor type 1B (Mel-1B-R) (Mel1b melatonin receptor). | |||||
|
OR8J3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | -0.003627 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGG0, Q96RC2 | Gene names | OR8J3 | |||
|
Domain Architecture |
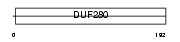
|
|||||
| Description | Olfactory receptor 8J3. | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.004443 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
YTHD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.005895 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
CEBPZ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.006689 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53569, Q3TXW5, Q3TYA8, Q3UWQ7 | Gene names | Cebpz, Cbf2, Cebpa-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
DCR1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.006523 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96SD1, Q5JSR4, Q5JSR5, Q5JSR7, Q5JSR8, Q5JSR9, Q5JSS0, Q5JSS7, Q6PK14, Q8N101, Q8N132, Q8TBW9, Q9BVW9, Q9HAM4 | Gene names | DCLRE1C, ARTEMIS, ASCID, SCIDA, SNM1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (SNM1- like protein) (A-SCID protein) (hSNM1C). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | -0.005662 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | -0.005700 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
OR8J1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | -0.003715 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 633 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGP2, Q96RC3 | Gene names | OR8J1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 8J1. | |||||
|
PLAL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | -0.003572 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UM63 | Gene names | PLAGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein PLAGL1 (Pleiomorphic adenoma-like protein 1). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | -0.003498 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.005133 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
ZF161_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | -0.004358 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43829, O00403 | Gene names | ZFP161, ZBTB14 | |||
|
Domain Architecture |
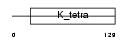
|
|||||
| Description | Zinc finger protein 161 homolog (Zfp-161) (Zinc finger protein 5) (hZF5) (Zinc finger and BTB domain-containing protein 14). | |||||
|
ZF161_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | -0.004359 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08376 | Gene names | Zfp161, Zfp-5, Zfp5 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 161 (Zfp-161) (Zinc finger protein 5) (ZF5). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.002902 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
LAX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.007265 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWV1, Q6NSZ6, Q9NXB4 | Gene names | LAX1, LAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte transmembrane adapter 1 (Membrane-associated adapter protein LAX) (Linker for activation of X cells). | |||||
|
ATP7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.136927 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.135326 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
ATP7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.140951 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.140998 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
AT10B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT10D_HUMAN
|
||||||
| NC score | 0.993605 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_MOUSE
|
||||||
| NC score | 0.992781 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10A_MOUSE
|
||||||
| NC score | 0.992165 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT10A_HUMAN
|
||||||
| NC score | 0.991708 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT8B4_HUMAN
|
||||||
| NC score | 0.950884 (rank : 6) | θ value | 2.36663e-117 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT8B2_HUMAN
|
||||||
| NC score | 0.950843 (rank : 7) | θ value | 1.81207e-117 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT8B1_HUMAN
|
||||||
| NC score | 0.947104 (rank : 8) | θ value | 1.69998e-107 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT8A2_MOUSE
|
||||||
| NC score | 0.945944 (rank : 9) | θ value | 3.77839e-115 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT8A2_HUMAN
|
||||||
| NC score | 0.945916 (rank : 10) | θ value | 2.44908e-114 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8A1_HUMAN
|
||||||
| NC score | 0.945433 (rank : 11) | θ value | 3.78717e-107 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_MOUSE
|
||||||
| NC score | 0.944993 (rank : 12) | θ value | 4.94618e-107 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8B3_HUMAN
|
||||||
| NC score | 0.943576 (rank : 13) | θ value | 4.96919e-91 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
AT11B_HUMAN
|
||||||
| NC score | 0.941849 (rank : 14) | θ value | 9.98939e-100 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT11C_HUMAN
|
||||||
| NC score | 0.940397 (rank : 15) | θ value | 3.80478e-91 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT11A_HUMAN
|
||||||
| NC score | 0.939810 (rank : 16) | θ value | 2.30293e-96 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11A_MOUSE
|
||||||
| NC score | 0.937357 (rank : 17) | θ value | 4.49445e-92 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
ATP9B_MOUSE
|
||||||
| NC score | 0.911951 (rank : 18) | θ value | 3.57772e-73 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_HUMAN
|
||||||
| NC score | 0.909303 (rank : 19) | θ value | 2.73936e-73 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9A_HUMAN
|
||||||
| NC score | 0.908914 (rank : 20) | θ value | 1.00825e-67 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
ATP9A_MOUSE
|
||||||
| NC score | 0.905638 (rank : 21) | θ value | 1.90147e-66 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
AT133_HUMAN
|
||||||
| NC score | 0.585031 (rank : 22) | θ value | 1.38499e-08 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT132_HUMAN
|
||||||
| NC score | 0.567380 (rank : 23) | θ value | 1.86753e-13 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT132_MOUSE
|
||||||
| NC score | 0.566926 (rank : 24) | θ value | 1.29331e-14 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT2B4_HUMAN
|
||||||
| NC score | 0.551641 (rank : 25) | θ value | 2.06002e-20 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.530326 (rank : 26) | θ value | 1.16975e-15 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.529355 (rank : 27) | θ value | 3.76295e-14 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| NC score | 0.525539 (rank : 28) | θ value | 6.41864e-14 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.502749 (rank : 29) | θ value | 1.16975e-15 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT131_MOUSE
|
||||||
| NC score | 0.457013 (rank : 30) | θ value | 5.44631e-05 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT131_HUMAN
|
||||||
| NC score | 0.455553 (rank : 31) | θ value | 3.19293e-05 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT12A_HUMAN
|
||||||
| NC score | 0.398982 (rank : 32) | θ value | 8.97725e-08 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT12A_MOUSE
|
||||||
| NC score | 0.395396 (rank : 33) | θ value | 3.41135e-07 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2C1_HUMAN
|
||||||
| NC score | 0.390128 (rank : 34) | θ value | 2.44474e-05 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2A2_MOUSE
|
||||||
| NC score | 0.388634 (rank : 35) | θ value | 0.00175202 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_HUMAN
|
||||||
| NC score | 0.387414 (rank : 36) | θ value | 0.00175202 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2C1_MOUSE
|
||||||
| NC score | 0.387377 (rank : 37) | θ value | 1.43324e-05 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2A3_HUMAN
|
||||||
| NC score | 0.381962 (rank : 38) | θ value | 0.0148317 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
ATP4A_MOUSE
|
||||||
| NC score | 0.380352 (rank : 39) | θ value | 1.43324e-05 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT1A4_HUMAN
|
||||||
| NC score | 0.376495 (rank : 40) | θ value | 2.88788e-06 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
ATP4A_HUMAN
|
||||||
| NC score | 0.375984 (rank : 41) | θ value | 1.43324e-05 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT1A4_MOUSE
|
||||||
| NC score | 0.375949 (rank : 42) | θ value | 4.92598e-06 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT2A1_MOUSE
|
||||||
| NC score | 0.374535 (rank : 43) | θ value | 0.00509761 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A3_MOUSE
|
||||||
| NC score | 0.372414 (rank : 44) | θ value | 0.0148317 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A1_HUMAN
|
||||||
| NC score | 0.372092 (rank : 45) | θ value | 0.00665767 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT1A1_MOUSE
|
||||||
| NC score | 0.370991 (rank : 46) | θ value | 9.29e-05 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A1_HUMAN
|
||||||
| NC score | 0.370363 (rank : 47) | θ value | 4.1701e-05 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A2_HUMAN
|
||||||
| NC score | 0.369385 (rank : 48) | θ value | 3.19293e-05 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| NC score | 0.368993 (rank : 49) | θ value | 3.19293e-05 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A3_HUMAN
|
||||||
| NC score | 0.350867 (rank : 50) | θ value | 0.000158464 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| NC score | 0.348456 (rank : 51) | θ value | 0.000158464 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.330920 (rank : 52) | θ value | 0.00228821 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
ATP7B_MOUSE
|
||||||
| NC score | 0.140998 (rank : 53) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
ATP7B_HUMAN
|
||||||
| NC score | 0.140951 (rank : 54) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7A_HUMAN
|
||||||
| NC score | 0.136927 (rank : 55) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| NC score | 0.135326 (rank : 56) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
PUM1_MOUSE
|
||||||
| NC score | 0.023605 (rank : 57) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U78, Q80X96, Q80YU8, Q8BPV7, Q9EPU6 | Gene names | Pum1, Kiaa0099 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1. | |||||
|
PUM1_HUMAN
|
||||||
| NC score | 0.023577 (rank : 58) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14671, Q5VXY7, Q9HAN1 | Gene names | PUM1, KIAA0099, PUMH1 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 1 (Pumilio-1) (HsPUM). | |||||
|
MMEL1_MOUSE
|
||||||
| NC score | 0.011499 (rank : 59) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLI3, Q3U495, Q9ERK2, Q9ERK3, Q9QZV6, Q9QZV7 | Gene names | Mmel1, Nep2, Nl1, Sep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) (Neprilysin-like peptidase) (NEPLP) (Neprilysin-like 1) (NL-1) (Soluble secreted endopeptidase) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
TSH1_HUMAN
|
||||||
| NC score | 0.011172 (rank : 60) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
RNF12_HUMAN
|
||||||
| NC score | 0.010205 (rank : 61) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.009675 (rank : 62) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
RD23B_HUMAN
|
||||||
| NC score | 0.008805 (rank : 63) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
LAX1_HUMAN
|
||||||
| NC score | 0.007265 (rank : 64) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWV1, Q6NSZ6, Q9NXB4 | Gene names | LAX1, LAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte transmembrane adapter 1 (Membrane-associated adapter protein LAX) (Linker for activation of X cells). | |||||
|
KI20A_HUMAN
|
||||||
| NC score | 0.006855 (rank : 65) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95235 | Gene names | KIF20A, RAB6KIFL | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin family member 20A (Rabkinesin-6) (Rab6-interacting kinesin- like protein) (GG10_2). | |||||
|
CEBPZ_MOUSE
|
||||||
| NC score | 0.006689 (rank : 66) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53569, Q3TXW5, Q3TYA8, Q3UWQ7 | Gene names | Cebpz, Cbf2, Cebpa-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
DCR1C_HUMAN
|
||||||
| NC score | 0.006523 (rank : 67) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96SD1, Q5JSR4, Q5JSR5, Q5JSR7, Q5JSR8, Q5JSR9, Q5JSS0, Q5JSS7, Q6PK14, Q8N101, Q8N132, Q8TBW9, Q9BVW9, Q9HAM4 | Gene names | DCLRE1C, ARTEMIS, ASCID, SCIDA, SNM1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (SNM1- like protein) (A-SCID protein) (hSNM1C). | |||||
|
YTHD1_HUMAN
|
||||||
| NC score | 0.005895 (rank : 68) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.005797 (rank : 69) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.005747 (rank : 70) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.005621 (rank : 71) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.005133 (rank : 72) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
S6A18_MOUSE
|
||||||
| NC score | 0.005037 (rank : 73) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88576, O88577, O88578, O88579, O88580, O88581, Q91XG6 | Gene names | Slc6a18, Xt2, Xtrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent transporter XTRP2 (Solute carrier family 6 member 18). | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.004443 (rank : 74) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
KIFC2_MOUSE
|
||||||
| NC score | 0.003811 (rank : 75) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08672, O08613 | Gene names | Kifc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC2. | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.002902 (rank : 76) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.002674 (rank : 77) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.000752 (rank : 78) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | -0.000065 (rank : 79) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
KINH_MOUSE
|
||||||
| NC score | -0.000299 (rank : 80) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MTR1B_HUMAN
|
||||||
| NC score | -0.003236 (rank : 81) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49286 | Gene names | MTNR1B | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin receptor type 1B (Mel-1B-R) (Mel1b melatonin receptor). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | -0.003498 (rank : 82) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
PLAL1_HUMAN
|
||||||
| NC score | -0.003572 (rank : 83) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UM63 | Gene names | PLAGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein PLAGL1 (Pleiomorphic adenoma-like protein 1). | |||||
|
OR8J3_HUMAN
|
||||||
| NC score | -0.003627 (rank : 84) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGG0, Q96RC2 | Gene names | OR8J3 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 8J3. | |||||
|
OR8J1_HUMAN
|
||||||
| NC score | -0.003715 (rank : 85) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 633 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGP2, Q96RC3 | Gene names | OR8J1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 8J1. | |||||
|
ZF161_HUMAN
|
||||||
| NC score | -0.004358 (rank : 86) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43829, O00403 | Gene names | ZFP161, ZBTB14 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 161 homolog (Zfp-161) (Zinc finger protein 5) (hZF5) (Zinc finger and BTB domain-containing protein 14). | |||||
|
ZF161_MOUSE
|
||||||
| NC score | -0.004359 (rank : 87) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08376 | Gene names | Zfp161, Zfp-5, Zfp5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 161 (Zfp-161) (Zinc finger protein 5) (ZF5). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | -0.005662 (rank : 88) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | -0.005700 (rank : 89) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||