Please be patient as the page loads
|
AT11A_HUMAN
|
||||||
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AT11A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998097 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.993232 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT11C_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.995118 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT8A1_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.977403 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.977422 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A2_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.976696 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8A2_MOUSE
|
||||||
| θ value | 1.71888e-184 (rank : 8) | NC score | 0.976869 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT8B4_HUMAN
|
||||||
| θ value | 5.1754e-181 (rank : 9) | NC score | 0.977858 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT8B2_HUMAN
|
||||||
| θ value | 1.66487e-179 (rank : 10) | NC score | 0.977909 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT8B1_HUMAN
|
||||||
| θ value | 7.49059e-172 (rank : 11) | NC score | 0.976869 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT8B3_HUMAN
|
||||||
| θ value | 5.96274e-153 (rank : 12) | NC score | 0.972821 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
ATP9A_HUMAN
|
||||||
| θ value | 4.02752e-125 (rank : 13) | NC score | 0.947765 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
ATP9A_MOUSE
|
||||||
| θ value | 2.88631e-123 (rank : 14) | NC score | 0.945177 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
ATP9B_HUMAN
|
||||||
| θ value | 2.06847e-121 (rank : 15) | NC score | 0.946247 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_MOUSE
|
||||||
| θ value | 7.6132e-116 (rank : 16) | NC score | 0.947615 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
AT10D_HUMAN
|
||||||
| θ value | 6.91785e-101 (rank : 17) | NC score | 0.934713 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_MOUSE
|
||||||
| θ value | 1.30465e-99 (rank : 18) | NC score | 0.934584 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10B_HUMAN
|
||||||
| θ value | 2.30293e-96 (rank : 19) | NC score | 0.939810 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT10A_MOUSE
|
||||||
| θ value | 1.06975e-93 (rank : 20) | NC score | 0.933015 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT10A_HUMAN
|
||||||
| θ value | 2.30828e-88 (rank : 21) | NC score | 0.930594 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT2B4_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 22) | NC score | 0.555992 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 23) | NC score | 0.535980 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 24) | NC score | 0.507181 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT2B2_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 25) | NC score | 0.530933 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 26) | NC score | 0.534517 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT12A_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 27) | NC score | 0.401184 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT132_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 28) | NC score | 0.532842 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT12A_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 29) | NC score | 0.397899 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT132_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 30) | NC score | 0.534890 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
ATP4A_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 31) | NC score | 0.389201 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
ATP4A_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 32) | NC score | 0.384311 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT133_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 33) | NC score | 0.570153 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT2A1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 34) | NC score | 0.387699 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 35) | NC score | 0.385340 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT1A1_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 36) | NC score | 0.369829 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A4_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 37) | NC score | 0.373896 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT131_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 38) | NC score | 0.458774 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT1A2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 39) | NC score | 0.368321 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT2A3_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 40) | NC score | 0.391171 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT131_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 41) | NC score | 0.454641 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT1A2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 42) | NC score | 0.368710 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A4_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 43) | NC score | 0.372068 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT1A1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 44) | NC score | 0.366853 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT2A2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 45) | NC score | 0.395188 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 46) | NC score | 0.396294 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT1A3_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 47) | NC score | 0.349319 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 48) | NC score | 0.346730 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2A3_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 49) | NC score | 0.382797 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2C1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.379368 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.375262 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.322601 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.006412 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
CRCM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.009248 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
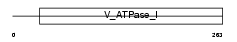
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.000539 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | -0.000765 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
T2R14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.009983 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYV8 | Gene names | TAS2R14 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 14 (T2R14) (Taste receptor family B member 1) (TRB1). | |||||
|
CTNL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.012400 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88327, Q810J3 | Gene names | Ctnnal1, Catnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-catulin (Catenin alpha-like protein 1) (Alpha-catenin-related protein) (ACRP). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.024185 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.024088 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
NARGL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.007167 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NOX5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.009265 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.020873 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.020638 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
VGFR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | -0.003477 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35916 | Gene names | FLT4 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 3 precursor (EC 2.7.10.1) (VEGFR-3) (Tyrosine-protein kinase receptor FLT4). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.020345 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.020222 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.001941 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.016413 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
CDK6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | -0.003556 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64261, Q9R1D2, Q9R1D3 | Gene names | Cdk6, Crk2 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 6 (EC 2.7.11.22) (Serine/threonine- protein kinase PLSTIRE) (CRK2). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.000260 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
SC6A7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.004279 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99884, Q52LU6 | Gene names | SLC6A7, PROT | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent proline transporter (Solute carrier family 6 member 7). | |||||
|
SC6A7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.004279 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGE7, Q80UM1 | Gene names | Slc6a7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent proline transporter (Solute carrier family 6 member 7). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.019447 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.019752 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
CDK6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | -0.003779 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00534 | Gene names | CDK6 | |||
|
Domain Architecture |
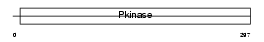
|
|||||
| Description | Cell division protein kinase 6 (EC 2.7.11.22) (Serine/threonine- protein kinase PLSTIRE). | |||||
|
CTNL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.009687 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBT7, O76084, Q53FQ2, Q5JTQ7, Q5JTQ8, Q9Y401 | Gene names | CTNNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-catulin (Catenin alpha-like protein 1) (Alpha-catenin-related protein) (ACRP). | |||||
|
PSMD6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.006711 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15008 | Gene names | PSMD6, KIAA0107, PFAAP4 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 6 (26S proteasome regulatory subunit S10) (p42A) (Proteasome regulatory particle subunit p44S10) (Phosphonoformate immuno-associated protein 4) (Breast cancer- associated protein SGA-113M). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | -0.000042 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | -0.001083 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
K0090_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.005095 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N766, Q14700, Q63HL0, Q63HL3, Q8NBH8 | Gene names | KIAA0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
K0286_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.006098 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14524, Q68DH0, Q6IQ25 | Gene names | KIAA0286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0286. | |||||
|
PSN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.006252 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49768, O95465, Q14762, Q15719, Q15720, Q96P33, Q9UIF0 | Gene names | PSEN1, AD3, PS1, PSNL1 | |||
|
Domain Architecture |

|
|||||
| Description | Presenilin-1 (EC 3.4.23.-) (PS-1) (Protein S182) [Contains: Presenilin-1 NTF subunit; Presenilin-1 CTF subunit; Presenilin-1 CTF12 (PS1-CTF12)]. | |||||
|
PSN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.006249 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49769, Q91WK6, Q9JLP9 | Gene names | Psen1, Ad3h, Psnl1 | |||
|
Domain Architecture |

|
|||||
| Description | Presenilin-1 (EC 3.4.23.-) (PS-1) (Protein S182) [Contains: Presenilin-1 NTF subunit; Presenilin-1 CTF subunit; Presenilin-1 CTF12 (PS1-CTF12)]. | |||||
|
ATP7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.134099 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.132693 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
ATP7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.137270 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.137628 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
AT11A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11A_MOUSE
|
||||||
| NC score | 0.998097 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11C_HUMAN
|
||||||
| NC score | 0.995118 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT11B_HUMAN
|
||||||
| NC score | 0.993232 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT8B2_HUMAN
|
||||||
| NC score | 0.977909 (rank : 5) | θ value | 1.66487e-179 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT8B4_HUMAN
|
||||||
| NC score | 0.977858 (rank : 6) | θ value | 5.1754e-181 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT8A1_MOUSE
|
||||||
| NC score | 0.977422 (rank : 7) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_HUMAN
|
||||||
| NC score | 0.977403 (rank : 8) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A2_MOUSE
|
||||||
| NC score | 0.976869 (rank : 9) | θ value | 1.71888e-184 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT8B1_HUMAN
|
||||||
| NC score | 0.976869 (rank : 10) | θ value | 7.49059e-172 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT8A2_HUMAN
|
||||||
| NC score | 0.976696 (rank : 11) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8B3_HUMAN
|
||||||
| NC score | 0.972821 (rank : 12) | θ value | 5.96274e-153 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
ATP9A_HUMAN
|
||||||
| NC score | 0.947765 (rank : 13) | θ value | 4.02752e-125 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
ATP9B_MOUSE
|
||||||
| NC score | 0.947615 (rank : 14) | θ value | 7.6132e-116 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_HUMAN
|
||||||
| NC score | 0.946247 (rank : 15) | θ value | 2.06847e-121 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9A_MOUSE
|
||||||
| NC score | 0.945177 (rank : 16) | θ value | 2.88631e-123 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
AT10B_HUMAN
|
||||||
| NC score | 0.939810 (rank : 17) | θ value | 2.30293e-96 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT10D_HUMAN
|
||||||
| NC score | 0.934713 (rank : 18) | θ value | 6.91785e-101 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_MOUSE
|
||||||
| NC score | 0.934584 (rank : 19) | θ value | 1.30465e-99 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10A_MOUSE
|
||||||
| NC score | 0.933015 (rank : 20) | θ value | 1.06975e-93 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT10A_HUMAN
|
||||||
| NC score | 0.930594 (rank : 21) | θ value | 2.30828e-88 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT133_HUMAN
|
||||||
| NC score | 0.570153 (rank : 22) | θ value | 1.17247e-07 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT2B4_HUMAN
|
||||||
| NC score | 0.555992 (rank : 23) | θ value | 4.01107e-24 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.535980 (rank : 24) | θ value | 9.24701e-21 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT132_MOUSE
|
||||||
| NC score | 0.534890 (rank : 25) | θ value | 1.38499e-08 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.534517 (rank : 26) | θ value | 1.058e-16 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT132_HUMAN
|
||||||
| NC score | 0.532842 (rank : 27) | θ value | 3.64472e-09 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT2B2_MOUSE
|
||||||
| NC score | 0.530933 (rank : 28) | θ value | 2.7842e-17 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.507181 (rank : 29) | θ value | 3.88503e-19 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT131_MOUSE
|
||||||
| NC score | 0.458774 (rank : 30) | θ value | 1.09739e-05 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT131_HUMAN
|
||||||
| NC score | 0.454641 (rank : 31) | θ value | 3.19293e-05 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT12A_HUMAN
|
||||||
| NC score | 0.401184 (rank : 32) | θ value | 4.30538e-10 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT12A_MOUSE
|
||||||
| NC score | 0.397899 (rank : 33) | θ value | 4.76016e-09 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2A2_MOUSE
|
||||||
| NC score | 0.396294 (rank : 34) | θ value | 0.000270298 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_HUMAN
|
||||||
| NC score | 0.395188 (rank : 35) | θ value | 0.000158464 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A3_HUMAN
|
||||||
| NC score | 0.391171 (rank : 36) | θ value | 2.44474e-05 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
ATP4A_MOUSE
|
||||||
| NC score | 0.389201 (rank : 37) | θ value | 1.38499e-08 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT2A1_MOUSE
|
||||||
| NC score | 0.387699 (rank : 38) | θ value | 1.69304e-06 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A1_HUMAN
|
||||||
| NC score | 0.385340 (rank : 39) | θ value | 3.77169e-06 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
ATP4A_HUMAN
|
||||||
| NC score | 0.384311 (rank : 40) | θ value | 3.08544e-08 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT2A3_MOUSE
|
||||||
| NC score | 0.382797 (rank : 41) | θ value | 0.00665767 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2C1_HUMAN
|
||||||
| NC score | 0.379368 (rank : 42) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C1_MOUSE
|
||||||
| NC score | 0.375262 (rank : 43) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT1A4_HUMAN
|
||||||
| NC score | 0.373896 (rank : 44) | θ value | 8.40245e-06 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT1A4_MOUSE
|
||||||
| NC score | 0.372068 (rank : 45) | θ value | 4.1701e-05 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT1A1_MOUSE
|
||||||
| NC score | 0.369829 (rank : 46) | θ value | 6.43352e-06 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A2_HUMAN
|
||||||
| NC score | 0.368710 (rank : 47) | θ value | 3.19293e-05 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| NC score | 0.368321 (rank : 48) | θ value | 2.44474e-05 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A1_HUMAN
|
||||||
| NC score | 0.366853 (rank : 49) | θ value | 0.000158464 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A3_HUMAN
|
||||||
| NC score | 0.349319 (rank : 50) | θ value | 0.000602161 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| NC score | 0.346730 (rank : 51) | θ value | 0.00134147 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.322601 (rank : 52) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
ATP7B_MOUSE
|
||||||
| NC score | 0.137628 (rank : 53) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
ATP7B_HUMAN
|
||||||
| NC score | 0.137270 (rank : 54) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7A_HUMAN
|
||||||
| NC score | 0.134099 (rank : 55) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| NC score | 0.132693 (rank : 56) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.024185 (rank : 57) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.024088 (rank : 58) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.020873 (rank : 59) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.020638 (rank : 60) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.020345 (rank : 61) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.020222 (rank : 62) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
TF7L2_MOUSE
|
||||||
| NC score | 0.019752 (rank : 63) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.019447 (rank : 64) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.016413 (rank : 65) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
CTNL1_MOUSE
|
||||||
| NC score | 0.012400 (rank : 66) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88327, Q810J3 | Gene names | Ctnnal1, Catnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-catulin (Catenin alpha-like protein 1) (Alpha-catenin-related protein) (ACRP). | |||||
|
T2R14_HUMAN
|
||||||
| NC score | 0.009983 (rank : 67) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYV8 | Gene names | TAS2R14 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 14 (T2R14) (Taste receptor family B member 1) (TRB1). | |||||
|
CTNL1_HUMAN
|
||||||
| NC score | 0.009687 (rank : 68) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBT7, O76084, Q53FQ2, Q5JTQ7, Q5JTQ8, Q9Y401 | Gene names | CTNNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-catulin (Catenin alpha-like protein 1) (Alpha-catenin-related protein) (ACRP). | |||||
|
NOX5_HUMAN
|
||||||
| NC score | 0.009265 (rank : 69) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.009248 (rank : 70) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
NARGL_HUMAN
|
||||||
| NC score | 0.007167 (rank : 71) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
PSMD6_HUMAN
|
||||||
| NC score | 0.006711 (rank : 72) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15008 | Gene names | PSMD6, KIAA0107, PFAAP4 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 6 (26S proteasome regulatory subunit S10) (p42A) (Proteasome regulatory particle subunit p44S10) (Phosphonoformate immuno-associated protein 4) (Breast cancer- associated protein SGA-113M). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.006412 (rank : 73) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
PSN1_HUMAN
|
||||||
| NC score | 0.006252 (rank : 74) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49768, O95465, Q14762, Q15719, Q15720, Q96P33, Q9UIF0 | Gene names | PSEN1, AD3, PS1, PSNL1 | |||
|
Domain Architecture |

|
|||||
| Description | Presenilin-1 (EC 3.4.23.-) (PS-1) (Protein S182) [Contains: Presenilin-1 NTF subunit; Presenilin-1 CTF subunit; Presenilin-1 CTF12 (PS1-CTF12)]. | |||||
|
PSN1_MOUSE
|
||||||
| NC score | 0.006249 (rank : 75) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49769, Q91WK6, Q9JLP9 | Gene names | Psen1, Ad3h, Psnl1 | |||
|
Domain Architecture |

|
|||||
| Description | Presenilin-1 (EC 3.4.23.-) (PS-1) (Protein S182) [Contains: Presenilin-1 NTF subunit; Presenilin-1 CTF subunit; Presenilin-1 CTF12 (PS1-CTF12)]. | |||||
|
K0286_HUMAN
|
||||||
| NC score | 0.006098 (rank : 76) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14524, Q68DH0, Q6IQ25 | Gene names | KIAA0286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0286. | |||||
|
K0090_HUMAN
|
||||||
| NC score | 0.005095 (rank : 77) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N766, Q14700, Q63HL0, Q63HL3, Q8NBH8 | Gene names | KIAA0090 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0090 precursor. | |||||
|
SC6A7_MOUSE
|
||||||
| NC score | 0.004279 (rank : 78) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGE7, Q80UM1 | Gene names | Slc6a7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent proline transporter (Solute carrier family 6 member 7). | |||||
|
SC6A7_HUMAN
|
||||||
| NC score | 0.004279 (rank : 79) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99884, Q52LU6 | Gene names | SLC6A7, PROT | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent proline transporter (Solute carrier family 6 member 7). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.001941 (rank : 80) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.000539 (rank : 81) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.000260 (rank : 82) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | -0.000042 (rank : 83) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | -0.000765 (rank : 84) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | -0.001083 (rank : 85) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
VGFR3_HUMAN
|
||||||
| NC score | -0.003477 (rank : 86) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35916 | Gene names | FLT4 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 3 precursor (EC 2.7.10.1) (VEGFR-3) (Tyrosine-protein kinase receptor FLT4). | |||||
|
CDK6_MOUSE
|
||||||
| NC score | -0.003556 (rank : 87) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64261, Q9R1D2, Q9R1D3 | Gene names | Cdk6, Crk2 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 6 (EC 2.7.11.22) (Serine/threonine- protein kinase PLSTIRE) (CRK2). | |||||
|
CDK6_HUMAN
|
||||||
| NC score | -0.003779 (rank : 88) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00534 | Gene names | CDK6 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 6 (EC 2.7.11.22) (Serine/threonine- protein kinase PLSTIRE). | |||||