Please be patient as the page loads
|
TF7L2_MOUSE
|
||||||
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TF7L2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994412 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 2.98686e-120 (rank : 3) | NC score | 0.980799 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 3.09092e-117 (rank : 4) | NC score | 0.979045 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 2.5344e-111 (rank : 5) | NC score | 0.968595 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 4.32305e-111 (rank : 6) | NC score | 0.971259 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 1.66964e-54 (rank : 7) | NC score | 0.922739 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 1.08223e-53 (rank : 8) | NC score | 0.935406 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 9) | NC score | 0.524010 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 10) | NC score | 0.557174 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 11) | NC score | 0.616426 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 12) | NC score | 0.611257 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 13) | NC score | 0.582755 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 14) | NC score | 0.581761 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX8_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 15) | NC score | 0.593206 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 16) | NC score | 0.600261 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 17) | NC score | 0.570474 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 18) | NC score | 0.567584 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX2_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 19) | NC score | 0.591195 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 20) | NC score | 0.590853 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 21) | NC score | 0.595939 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX8_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 22) | NC score | 0.589908 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 23) | NC score | 0.578198 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 24) | NC score | 0.580048 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 25) | NC score | 0.574113 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 26) | NC score | 0.573902 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 27) | NC score | 0.574458 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX10_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 28) | NC score | 0.572072 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 29) | NC score | 0.593786 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 30) | NC score | 0.593844 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SRY_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 31) | NC score | 0.608423 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX12_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 32) | NC score | 0.608650 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX12_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 33) | NC score | 0.610009 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SRY_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 34) | NC score | 0.595742 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 35) | NC score | 0.584771 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX17_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 36) | NC score | 0.576672 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 37) | NC score | 0.582831 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX17_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 38) | NC score | 0.578996 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 39) | NC score | 0.591936 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 40) | NC score | 0.591898 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX18_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 41) | NC score | 0.580292 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX11_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 42) | NC score | 0.591110 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX11_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 43) | NC score | 0.589498 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
BBX_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 44) | NC score | 0.489996 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 45) | NC score | 0.493172 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
SOX1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 46) | NC score | 0.574606 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 47) | NC score | 0.574147 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX3_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 48) | NC score | 0.560720 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX3_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 49) | NC score | 0.563840 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX18_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 50) | NC score | 0.573578 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
HBP1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 51) | NC score | 0.510958 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 52) | NC score | 0.512479 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HM20A_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 53) | NC score | 0.351225 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 54) | NC score | 0.350345 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
SOX15_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 55) | NC score | 0.561154 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX15_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 56) | NC score | 0.566026 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 57) | NC score | 0.304124 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 58) | NC score | 0.292747 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HM20B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.343845 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
HM20B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.347678 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.025687 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.032261 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.317208 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.320686 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.022353 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.021524 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.017555 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.026426 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.183531 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
AMOL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.013271 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.022607 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.016650 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
TFAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.192771 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.011722 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.008725 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.002710 (rank : 113) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.023684 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TNR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.022375 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |
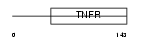
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
AT11A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.019752 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | -0.001209 (rank : 115) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.018188 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.028987 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.008458 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
FINC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.007253 (rank : 109) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.007540 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.267094 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.014709 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.013420 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.014282 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.002967 (rank : 112) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
AT11A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.016418 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
ATS20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.000349 (rank : 114) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.022252 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.005450 (rank : 110) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.003237 (rank : 111) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
CN092_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.132471 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.128051 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
GCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.104713 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.225675 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
HMG1X_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.194625 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
HMGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.243634 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.243718 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.246637 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.261525 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.258514 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.140481 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.073972 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.066385 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.063101 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.162232 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.152460 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SP100_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.104402 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.125867 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
TOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.098198 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.098304 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TF7L2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.994412 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.980799 (rank : 3) | θ value | 2.98686e-120 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.979045 (rank : 4) | θ value | 3.09092e-117 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.971259 (rank : 5) | θ value | 4.32305e-111 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.968595 (rank : 6) | θ value | 2.5344e-111 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.935406 (rank : 7) | θ value | 1.08223e-53 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.922739 (rank : 8) | θ value | 1.66964e-54 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.616426 (rank : 9) | θ value | 2.98157e-11 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.611257 (rank : 10) | θ value | 4.30538e-10 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX12_MOUSE
|
||||||
| NC score | 0.610009 (rank : 11) | θ value | 1.29631e-06 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.608650 (rank : 12) | θ value | 1.29631e-06 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SRY_HUMAN
|
||||||
| NC score | 0.608423 (rank : 13) | θ value | 9.92553e-07 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.600261 (rank : 14) | θ value | 5.26297e-08 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.595939 (rank : 15) | θ value | 6.87365e-08 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SRY_MOUSE
|
||||||
| NC score | 0.595742 (rank : 16) | θ value | 2.21117e-06 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX14_MOUSE
|
||||||
| NC score | 0.593844 (rank : 17) | θ value | 7.59969e-07 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.593786 (rank : 18) | θ value | 7.59969e-07 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX8_HUMAN
|
||||||
| NC score | 0.593206 (rank : 19) | θ value | 1.38499e-08 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.591936 (rank : 20) | θ value | 1.09739e-05 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.591898 (rank : 21) | θ value | 1.09739e-05 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX2_HUMAN
|
||||||
| NC score | 0.591195 (rank : 22) | θ value | 6.87365e-08 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX11_HUMAN
|
||||||
| NC score | 0.591110 (rank : 23) | θ value | 2.44474e-05 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX2_MOUSE
|
||||||
| NC score | 0.590853 (rank : 24) | θ value | 6.87365e-08 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.589908 (rank : 25) | θ value | 8.97725e-08 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX11_MOUSE
|
||||||
| NC score | 0.589498 (rank : 26) | θ value | 2.44474e-05 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.584771 (rank : 27) | θ value | 2.88788e-06 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.582831 (rank : 28) | θ value | 4.92598e-06 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.582755 (rank : 29) | θ value | 1.38499e-08 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.581761 (rank : 30) | θ value | 1.38499e-08 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX18_MOUSE
|
||||||
| NC score | 0.580292 (rank : 31) | θ value | 1.87187e-05 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.580048 (rank : 32) | θ value | 1.17247e-07 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX17_HUMAN
|
||||||
| NC score | 0.578996 (rank : 33) | θ value | 1.09739e-05 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.578198 (rank : 34) | θ value | 1.17247e-07 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX17_MOUSE
|
||||||
| NC score | 0.576672 (rank : 35) | θ value | 3.77169e-06 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX1_HUMAN
|
||||||
| NC score | 0.574606 (rank : 36) | θ value | 7.1131e-05 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.574458 (rank : 37) | θ value | 4.45536e-07 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX1_MOUSE
|
||||||
| NC score | 0.574147 (rank : 38) | θ value | 7.1131e-05 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.574113 (rank : 39) | θ value | 4.45536e-07 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.573902 (rank : 40) | θ value | 4.45536e-07 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX18_HUMAN
|
||||||
| NC score | 0.573578 (rank : 41) | θ value | 0.000270298 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.572072 (rank : 42) | θ value | 5.81887e-07 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.570474 (rank : 43) | θ value | 6.87365e-08 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.567584 (rank : 44) | θ value | 6.87365e-08 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX15_MOUSE
|
||||||
| NC score | 0.566026 (rank : 45) | θ value | 0.000786445 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.563840 (rank : 46) | θ value | 0.00020696 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX15_HUMAN
|
||||||
| NC score | 0.561154 (rank : 47) | θ value | 0.000786445 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.560720 (rank : 48) | θ value | 0.00020696 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.557174 (rank : 49) | θ value | 4.59992e-12 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.524010 (rank : 50) | θ value | 4.59992e-12 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HBP1_MOUSE
|
||||||
| NC score | 0.512479 (rank : 51) | θ value | 0.00035302 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_HUMAN
|
||||||
| NC score | 0.510958 (rank : 52) | θ value | 0.00035302 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.493172 (rank : 53) | θ value | 5.44631e-05 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_HUMAN
|
||||||
| NC score | 0.489996 (rank : 54) | θ value | 5.44631e-05 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.351225 (rank : 55) | θ value | 0.000602161 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.350345 (rank : 56) | θ value | 0.000602161 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
HM20B_HUMAN
|
||||||
| NC score | 0.347678 (rank : 57) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20B_MOUSE
|
||||||
| NC score | 0.343845 (rank : 58) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.320686 (rank : 59) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.317208 (rank : 60) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.304124 (rank : 61) | θ value | 0.0961366 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.292747 (rank : 62) | θ value | 0.163984 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.267094 (rank : 63) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB3_HUMAN
|
||||||
| NC score | 0.261525 (rank : 64) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_MOUSE
|
||||||
| NC score | 0.258514 (rank : 65) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.246637 (rank : 66) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB1_MOUSE
|
||||||
| NC score | 0.243718 (rank : 67) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_HUMAN
|
||||||
| NC score | 0.243634 (rank : 68) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.225675 (rank : 69) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
HMG1X_HUMAN
|
||||||
| NC score | 0.194625 (rank : 70) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
TFAM_MOUSE
|
||||||
| NC score | 0.192771 (rank : 71) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.183531 (rank : 72) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.162232 (rank : 73) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.152460 (rank : 74) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.140481 (rank : 75) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.132471 (rank : 76) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.128051 (rank : 77) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.125867 (rank : 78) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.104713 (rank : 79) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.104402 (rank : 80) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.098304 (rank : 81) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.098198 (rank : 82) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.073972 (rank : 83) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.066385 (rank : 84) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.063101 (rank : 85) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.032261 (rank : 86) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.028987 (rank : 87) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.026426 (rank : 88) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.025687 (rank : 89) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.023684 (rank : 90) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.022607 (rank : 91) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
TNR3_HUMAN
|
||||||
| NC score | 0.022375 (rank : 92) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.022353 (rank : 93) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.022252 (rank : 94) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.021524 (rank : 95) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
AT11A_HUMAN
|
||||||
| NC score | 0.019752 (rank : 96) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.018188 (rank : 97) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.017555 (rank : 98) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.016650 (rank : 99) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
AT11A_MOUSE
|
||||||
| NC score | 0.016418 (rank : 100) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.014709 (rank : 101) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.014282 (rank : 102) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.013420 (rank : 103) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
AMOL1_HUMAN
|
||||||
| NC score | 0.013271 (rank : 104) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.011722 (rank : 105) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.008725 (rank : 106) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.008458 (rank : 107) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.007540 (rank : 108) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.007253 (rank : 109) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.005450 (rank : 110) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.003237 (rank : 111) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.002967 (rank : 112) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.002710 (rank : 113) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
ATS20_MOUSE
|
||||||
| NC score | 0.000349 (rank : 114) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | -0.001209 (rank : 115) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||