Please be patient as the page loads
|
SOX10_HUMAN
|
||||||
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SOX10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX10_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998743 (rank : 2) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 1.93155e-127 (rank : 3) | NC score | 0.978381 (rank : 5) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 1.25199e-126 (rank : 4) | NC score | 0.976263 (rank : 6) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX8_MOUSE
|
||||||
| θ value | 3.22093e-90 (rank : 5) | NC score | 0.982259 (rank : 4) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX8_HUMAN
|
||||||
| θ value | 1.59853e-89 (rank : 6) | NC score | 0.982453 (rank : 3) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX2_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 7) | NC score | 0.908172 (rank : 26) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX12_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 8) | NC score | 0.940826 (rank : 8) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX12_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 9) | NC score | 0.943592 (rank : 7) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX2_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 10) | NC score | 0.907791 (rank : 27) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 11) | NC score | 0.923023 (rank : 14) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX17_MOUSE
|
||||||
| θ value | 7.06379e-29 (rank : 12) | NC score | 0.899877 (rank : 31) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX17_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 13) | NC score | 0.912127 (rank : 24) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 14) | NC score | 0.902447 (rank : 29) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 15) | NC score | 0.924210 (rank : 13) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 2.27234e-27 (rank : 16) | NC score | 0.924235 (rank : 12) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 17) | NC score | 0.913219 (rank : 22) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 18) | NC score | 0.913294 (rank : 21) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 19) | NC score | 0.924529 (rank : 11) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX18_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 20) | NC score | 0.917541 (rank : 17) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX18_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 21) | NC score | 0.917377 (rank : 18) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 22) | NC score | 0.895674 (rank : 32) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX3_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 23) | NC score | 0.909991 (rank : 25) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX1_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 24) | NC score | 0.913424 (rank : 20) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX11_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 25) | NC score | 0.924564 (rank : 10) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX15_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 26) | NC score | 0.918933 (rank : 16) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX1_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 27) | NC score | 0.912145 (rank : 23) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX11_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 28) | NC score | 0.927408 (rank : 9) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX3_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 29) | NC score | 0.905483 (rank : 28) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX15_MOUSE
|
||||||
| θ value | 2.77775e-25 (rank : 30) | NC score | 0.922311 (rank : 15) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SRY_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 31) | NC score | 0.900121 (rank : 30) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 32) | NC score | 0.817981 (rank : 34) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 33) | NC score | 0.820487 (rank : 33) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 34) | NC score | 0.792203 (rank : 38) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 35) | NC score | 0.794882 (rank : 37) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 36) | NC score | 0.800288 (rank : 35) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 37) | NC score | 0.799238 (rank : 36) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SRY_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 38) | NC score | 0.914030 (rank : 19) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 39) | NC score | 0.787053 (rank : 40) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 40) | NC score | 0.788350 (rank : 39) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 41) | NC score | 0.634396 (rank : 43) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 42) | NC score | 0.597502 (rank : 48) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 43) | NC score | 0.561082 (rank : 55) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
HMGB3_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 44) | NC score | 0.605544 (rank : 47) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 45) | NC score | 0.563491 (rank : 54) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 46) | NC score | 0.566118 (rank : 53) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
HMGB3_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 47) | NC score | 0.594958 (rank : 49) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 48) | NC score | 0.559798 (rank : 57) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 49) | NC score | 0.570821 (rank : 52) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L2_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 50) | NC score | 0.574113 (rank : 51) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
BBX_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 51) | NC score | 0.620782 (rank : 44) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 52) | NC score | 0.578913 (rank : 50) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
HBP1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 53) | NC score | 0.658976 (rank : 42) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 54) | NC score | 0.659169 (rank : 41) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 55) | NC score | 0.546262 (rank : 59) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 56) | NC score | 0.534730 (rank : 60) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 57) | NC score | 0.507600 (rank : 62) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 58) | NC score | 0.507646 (rank : 61) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 59) | NC score | 0.609989 (rank : 46) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
HMG1X_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 60) | NC score | 0.477095 (rank : 63) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 61) | NC score | 0.559922 (rank : 56) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 62) | NC score | 0.612041 (rank : 45) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 63) | NC score | 0.558773 (rank : 58) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 64) | NC score | 0.450340 (rank : 66) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 65) | NC score | 0.442711 (rank : 68) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 66) | NC score | 0.444873 (rank : 67) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
SP100_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 67) | NC score | 0.286389 (rank : 73) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 68) | NC score | 0.263694 (rank : 75) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 69) | NC score | 0.043367 (rank : 85) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 70) | NC score | 0.041569 (rank : 86) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 0.163984 (rank : 71) | NC score | 0.051007 (rank : 83) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 72) | NC score | 0.038880 (rank : 87) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 73) | NC score | 0.301957 (rank : 72) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 74) | NC score | 0.283945 (rank : 74) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 75) | NC score | 0.024457 (rank : 95) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
SENP5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.025933 (rank : 93) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
TFAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.315197 (rank : 71) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | -0.003900 (rank : 132) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.005799 (rank : 120) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
MED12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.015698 (rank : 100) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.043826 (rank : 84) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.241884 (rank : 77) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
SCG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.013448 (rank : 103) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.024511 (rank : 94) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.003923 (rank : 125) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.007935 (rank : 113) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.025995 (rank : 92) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.011057 (rank : 105) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.031747 (rank : 90) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.006251 (rank : 118) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.010586 (rank : 106) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.004211 (rank : 123) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.023497 (rank : 96) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
CDX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.000715 (rank : 128) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |
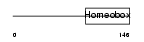
|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.001221 (rank : 127) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.008100 (rank : 112) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.006022 (rank : 119) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.008799 (rank : 110) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NAGPA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.004420 (rank : 122) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.013750 (rank : 102) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PEPP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.000570 (rank : 129) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQY4, Q9BR00 | Gene names | PEPP2, THG1 | |||
|
Domain Architecture |

|
|||||
| Description | Paired-like homeobox protein PEPP-2 (Testis homeobox gene 1). | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.009715 (rank : 109) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | -0.000910 (rank : 131) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | -0.000766 (rank : 130) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.035172 (rank : 88) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.026729 (rank : 91) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.007048 (rank : 116) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FA48A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.008131 (rank : 111) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TT00, Q8BG53, Q9JLS9 | Gene names | Fam48a, D3Ertd300e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
GLE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.002282 (rank : 126) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
GP156_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.010032 (rank : 108) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.015945 (rank : 99) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
LEG3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.010298 (rank : 107) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |
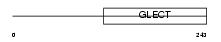
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.005782 (rank : 121) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
ZYX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.006469 (rank : 117) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.033997 (rank : 89) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CO039_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.014862 (rank : 101) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.019827 (rank : 98) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.022239 (rank : 97) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SC24B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.007593 (rank : 115) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
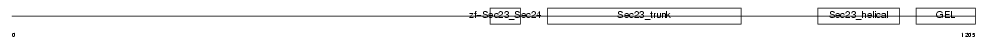
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.013336 (rank : 104) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SRC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.003925 (rank : 124) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
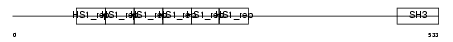
|
|||||
| Description | Src substrate cortactin. | |||||
|
TNR25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.007670 (rank : 114) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
CN092_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.191408 (rank : 78) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.185079 (rank : 79) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
GCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.138278 (rank : 82) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
HM20A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.381047 (rank : 69) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.379448 (rank : 70) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
HM20B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.471061 (rank : 64) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.450976 (rank : 65) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.253516 (rank : 76) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
TOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.166856 (rank : 81) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.167860 (rank : 80) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.998743 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX8_HUMAN
|
||||||
| NC score | 0.982453 (rank : 3) | θ value | 1.59853e-89 (rank : 6) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.982259 (rank : 4) | θ value | 3.22093e-90 (rank : 5) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.978381 (rank : 5) | θ value | 1.93155e-127 (rank : 3) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.976263 (rank : 6) | θ value | 1.25199e-126 (rank : 4) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX12_MOUSE
|
||||||
| NC score | 0.943592 (rank : 7) | θ value | 3.17079e-29 (rank : 9) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.940826 (rank : 8) | θ value | 3.17079e-29 (rank : 8) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX11_HUMAN
|
||||||
| NC score | 0.927408 (rank : 9) | θ value | 1.24688e-25 (rank : 28) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX11_MOUSE
|
||||||
| NC score | 0.924564 (rank : 10) | θ value | 5.59698e-26 (rank : 25) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.924529 (rank : 11) | θ value | 5.06226e-27 (rank : 19) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.924235 (rank : 12) | θ value | 2.27234e-27 (rank : 16) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.924210 (rank : 13) | θ value | 2.27234e-27 (rank : 15) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.923023 (rank : 14) | θ value | 5.40856e-29 (rank : 11) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX15_MOUSE
|
||||||
| NC score | 0.922311 (rank : 15) | θ value | 2.77775e-25 (rank : 30) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX15_HUMAN
|
||||||
| NC score | 0.918933 (rank : 16) | θ value | 5.59698e-26 (rank : 26) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX18_HUMAN
|
||||||
| NC score | 0.917541 (rank : 17) | θ value | 6.61148e-27 (rank : 20) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX18_MOUSE
|
||||||
| NC score | 0.917377 (rank : 18) | θ value | 1.92365e-26 (rank : 21) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SRY_HUMAN
|
||||||
| NC score | 0.914030 (rank : 19) | θ value | 2.27762e-19 (rank : 38) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX1_HUMAN
|
||||||
| NC score | 0.913424 (rank : 20) | θ value | 4.28545e-26 (rank : 24) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX14_MOUSE
|
||||||
| NC score | 0.913294 (rank : 21) | θ value | 5.06226e-27 (rank : 18) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.913219 (rank : 22) | θ value | 5.06226e-27 (rank : 17) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX1_MOUSE
|
||||||
| NC score | 0.912145 (rank : 23) | θ value | 7.30988e-26 (rank : 27) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX17_HUMAN
|
||||||
| NC score | 0.912127 (rank : 24) | θ value | 2.05525e-28 (rank : 13) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.909991 (rank : 25) | θ value | 3.28125e-26 (rank : 23) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX2_HUMAN
|
||||||
| NC score | 0.908172 (rank : 26) | θ value | 1.08979e-29 (rank : 7) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_MOUSE
|
||||||
| NC score | 0.907791 (rank : 27) | θ value | 4.14116e-29 (rank : 10) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.905483 (rank : 28) | θ value | 1.62847e-25 (rank : 29) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.902447 (rank : 29) | θ value | 1.02001e-27 (rank : 14) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SRY_MOUSE
|
||||||
| NC score | 0.900121 (rank : 30) | θ value | 6.40375e-22 (rank : 31) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX17_MOUSE
|
||||||
| NC score | 0.899877 (rank : 31) | θ value | 7.06379e-29 (rank : 12) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.895674 (rank : 32) | θ value | 2.51237e-26 (rank : 22) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.820487 (rank : 33) | θ value | 5.42112e-21 (rank : 33) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.817981 (rank : 34) | θ value | 5.42112e-21 (rank : 32) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.800288 (rank : 35) | θ value | 3.51386e-20 (rank : 36) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.799238 (rank : 36) | θ value | 3.51386e-20 (rank : 37) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.794882 (rank : 37) | θ value | 3.51386e-20 (rank : 35) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.792203 (rank : 38) | θ value | 3.51386e-20 (rank : 34) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.788350 (rank : 39) | θ value | 1.16975e-15 (rank : 40) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.787053 (rank : 40) | θ value | 1.16975e-15 (rank : 39) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
HBP1_MOUSE
|
||||||
| NC score | 0.659169 (rank : 41) | θ value | 2.21117e-06 (rank : 54) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_HUMAN
|
||||||
| NC score | 0.658976 (rank : 42) | θ value | 2.21117e-06 (rank : 53) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.634396 (rank : 43) | θ value | 8.67504e-11 (rank : 41) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BBX_HUMAN
|
||||||
| NC score | 0.620782 (rank : 44) | θ value | 1.69304e-06 (rank : 51) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.612041 (rank : 45) | θ value | 1.43324e-05 (rank : 62) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.609989 (rank : 46) | θ value | 1.09739e-05 (rank : 59) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
HMGB3_HUMAN
|
||||||
| NC score | 0.605544 (rank : 47) | θ value | 1.80886e-08 (rank : 44) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.597502 (rank : 48) | θ value | 1.47974e-10 (rank : 42) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HMGB3_MOUSE
|
||||||
| NC score | 0.594958 (rank : 49) | θ value | 1.99992e-07 (rank : 47) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.578913 (rank : 50) | θ value | 1.69304e-06 (rank : 52) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TF7L2_MOUSE
|
||||||
| NC score | 0.574113 (rank : 51) | θ value | 4.45536e-07 (rank : 50) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.570821 (rank : 52) | θ value | 4.45536e-07 (rank : 49) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.566118 (rank : 53) | θ value | 1.17247e-07 (rank : 46) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.563491 (rank : 54) | θ value | 2.36244e-08 (rank : 45) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.561082 (rank : 55) | θ value | 8.11959e-09 (rank : 43) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.559922 (rank : 56) | θ value | 1.43324e-05 (rank : 61) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.559798 (rank : 57) | θ value | 2.61198e-07 (rank : 48) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.558773 (rank : 58) | θ value | 1.87187e-05 (rank : 63) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.546262 (rank : 59) | θ value | 2.21117e-06 (rank : 55) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.534730 (rank : 60) | θ value | 4.92598e-06 (rank : 56) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB1_MOUSE
|
||||||
| NC score | 0.507646 (rank : 61) | θ value | 6.43352e-06 (rank : 58) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_HUMAN
|
||||||
| NC score | 0.507600 (rank : 62) | θ value | 6.43352e-06 (rank : 57) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMG1X_HUMAN
|
||||||
| NC score | 0.477095 (rank : 63) | θ value | 1.43324e-05 (rank : 60) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
HM20B_HUMAN
|
||||||
| NC score | 0.471061 (rank : 64) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20B_MOUSE
|
||||||
| NC score | 0.450976 (rank : 65) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.450340 (rank : 66) | θ value | 0.000602161 (rank : 64) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.444873 (rank : 67) | θ value | 0.00869519 (rank : 66) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.442711 (rank : 68) | θ value | 0.000786445 (rank : 65) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.381047 (rank : 69) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.379448 (rank : 70) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
TFAM_MOUSE
|
||||||
| NC score | 0.315197 (rank : 71) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.301957 (rank : 72) | θ value | 0.279714 (rank : 73) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.286389 (rank : 73) | θ value | 0.00869519 (rank : 67) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.283945 (rank : 74) | θ value | 0.279714 (rank : 74) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.263694 (rank : 75) | θ value | 0.0431538 (rank : 68) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.253516 (rank : 76) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.241884 (rank : 77) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.191408 (rank : 78) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.185079 (rank : 79) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.167860 (rank : 80) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.166856 (rank : 81) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.138278 (rank : 82) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.051007 (rank : 83) | θ value | 0.163984 (rank : 71) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.043826 (rank : 84) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.043367 (rank : 85) | θ value | 0.0563607 (rank : 69) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.041569 (rank : 86) | θ value | 0.125558 (rank : 70) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.038880 (rank : 87) | θ value | 0.21417 (rank : 72) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.035172 (rank : 88) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.033997 (rank : 89) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.031747 (rank : 90) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.026729 (rank : 91) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.025995 (rank : 92) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SENP5_HUMAN
|
||||||
| NC score | 0.025933 (rank : 93) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.024511 (rank : 94) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.024457 (rank : 95) | θ value | 0.279714 (rank : 75) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.023497 (rank : 96) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.022239 (rank : 97) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.019827 (rank : 98) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.015945 (rank : 99) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.015698 (rank : 100) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.014862 (rank : 101) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.013750 (rank : 102) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SCG1_MOUSE
|
||||||
| NC score | 0.013448 (rank : 103) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.013336 (rank : 104) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.011057 (rank : 105) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.010586 (rank : 106) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LEG3_HUMAN
|
||||||
| NC score | 0.010298 (rank : 107) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
GP156_MOUSE
|
||||||
| NC score | 0.010032 (rank : 108) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.009715 (rank : 109) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.008799 (rank : 110) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
FA48A_MOUSE
|
||||||
| NC score | 0.008131 (rank : 111) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TT00, Q8BG53, Q9JLS9 | Gene names | Fam48a, D3Ertd300e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.008100 (rank : 112) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.007935 (rank : 113) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.007670 (rank : 114) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
SC24B_HUMAN
|
||||||
| NC score | 0.007593 (rank : 115) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
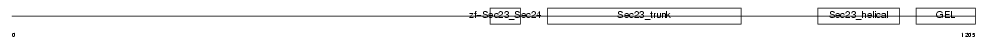
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.007048 (rank : 116) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ZYX_MOUSE
|
||||||
| NC score | 0.006469 (rank : 117) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.006251 (rank : 118) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.006022 (rank : 119) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.005799 (rank : 120) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.005782 (rank : 121) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
NAGPA_HUMAN
|
||||||
| NC score | 0.004420 (rank : 122) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.004211 (rank : 123) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SRC8_MOUSE
|
||||||
| NC score | 0.003925 (rank : 124) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
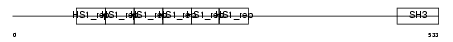
|
|||||
| Description | Src substrate cortactin. | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.003923 (rank : 125) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
GLE1_MOUSE
|
||||||
| NC score | 0.002282 (rank : 126) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.001221 (rank : 127) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
CDX1_HUMAN
|
||||||
| NC score | 0.000715 (rank : 128) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
PEPP2_HUMAN
|
||||||
| NC score | 0.000570 (rank : 129) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQY4, Q9BR00 | Gene names | PEPP2, THG1 | |||
|
Domain Architecture |

|
|||||
| Description | Paired-like homeobox protein PEPP-2 (Testis homeobox gene 1). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | -0.000766 (rank : 130) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | -0.000910 (rank : 131) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | -0.003900 (rank : 132) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||