Please be patient as the page loads
|
NCOA3_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NCOA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.934324 (rank : 3) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.932488 (rank : 4) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 171 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.966968 (rank : 2) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 3.5201e-129 (rank : 5) | NC score | 0.910960 (rank : 6) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 2.36115e-125 (rank : 6) | NC score | 0.912004 (rank : 5) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 7) | NC score | 0.603881 (rank : 9) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 8) | NC score | 0.604957 (rank : 8) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 9) | NC score | 0.575044 (rank : 13) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 10) | NC score | 0.574792 (rank : 14) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 11) | NC score | 0.605117 (rank : 7) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 12) | NC score | 0.602801 (rank : 10) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 13) | NC score | 0.563631 (rank : 21) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 14) | NC score | 0.555782 (rank : 25) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 15) | NC score | 0.582022 (rank : 12) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 16) | NC score | 0.589777 (rank : 11) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 17) | NC score | 0.569862 (rank : 16) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 18) | NC score | 0.566111 (rank : 19) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 19) | NC score | 0.566854 (rank : 18) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 20) | NC score | 0.565466 (rank : 20) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 21) | NC score | 0.555783 (rank : 24) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 22) | NC score | 0.562303 (rank : 22) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 23) | NC score | 0.561427 (rank : 23) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 24) | NC score | 0.572833 (rank : 15) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 25) | NC score | 0.569420 (rank : 17) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 26) | NC score | 0.553234 (rank : 26) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 27) | NC score | 0.537310 (rank : 27) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 28) | NC score | 0.527169 (rank : 28) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 29) | NC score | 0.521640 (rank : 30) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 30) | NC score | 0.524212 (rank : 29) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 31) | NC score | 0.074741 (rank : 43) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 32) | NC score | 0.051073 (rank : 78) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 33) | NC score | 0.055896 (rank : 64) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 34) | NC score | 0.057454 (rank : 58) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
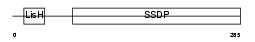
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 35) | NC score | 0.057454 (rank : 59) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
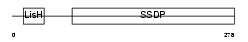
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
TEX15_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 36) | NC score | 0.059380 (rank : 56) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 37) | NC score | 0.055581 (rank : 66) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 38) | NC score | 0.364593 (rank : 31) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 39) | NC score | 0.357377 (rank : 32) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 40) | NC score | 0.007541 (rank : 161) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 41) | NC score | 0.084330 (rank : 41) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.064119 (rank : 52) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.115727 (rank : 40) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.032768 (rank : 99) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.007137 (rank : 163) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.009446 (rank : 147) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.025280 (rank : 112) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 48) | NC score | 0.049176 (rank : 83) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.006732 (rank : 165) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.019265 (rank : 125) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
HORN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.036146 (rank : 94) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.055713 (rank : 65) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SUHW4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.008510 (rank : 152) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.049435 (rank : 81) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.117686 (rank : 39) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TAOK1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.005367 (rank : 173) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 57) | NC score | 0.045412 (rank : 88) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.010482 (rank : 144) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.017641 (rank : 129) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.047011 (rank : 86) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.017404 (rank : 130) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.046753 (rank : 87) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.024046 (rank : 117) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.047391 (rank : 84) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 0.47712 (rank : 65) | NC score | 0.047161 (rank : 85) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
TAOK1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 66) | NC score | 0.005056 (rank : 178) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
N4BP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.033380 (rank : 97) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.031964 (rank : 101) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.032019 (rank : 100) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.039918 (rank : 91) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.035474 (rank : 96) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.028485 (rank : 106) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
SSXT_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.069481 (rank : 46) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |
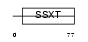
|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
MAML1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.052500 (rank : 71) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
SSXT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.067858 (rank : 47) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |
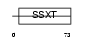
|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
ZBT46_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.004004 (rank : 180) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BID6, Q3U2R2, Q9D0G6, Q9D492 | Gene names | Zbtb46, Btbd4 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 46 (BTB/POZ domain- containing protein 4). | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.017117 (rank : 131) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.030909 (rank : 103) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
INVO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.022716 (rank : 119) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
K0240_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.029246 (rank : 105) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.055532 (rank : 67) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.024470 (rank : 114) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PCDA5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.003587 (rank : 183) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.031350 (rank : 102) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.041869 (rank : 90) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.006530 (rank : 166) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
FUS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.056318 (rank : 62) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
PCDA7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.005096 (rank : 177) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PRD16_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.000772 (rank : 191) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.013659 (rank : 135) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
ABCBB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.007416 (rank : 162) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95342, Q9UNB2 | Gene names | ABCB11, BSEP | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt export pump (ATP-binding cassette sub-family B member 11). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.008686 (rank : 151) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.032960 (rank : 98) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.000199 (rank : 193) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.012038 (rank : 142) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
INP5E_MOUSE
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.013545 (rank : 136) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.035920 (rank : 95) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.005469 (rank : 172) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
SNX15_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.024819 (rank : 113) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.008148 (rank : 153) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.037597 (rank : 92) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.007060 (rank : 164) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.007935 (rank : 155) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.003151 (rank : 186) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TTLL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.012444 (rank : 137) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
USBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.019767 (rank : 124) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
WT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | -0.000537 (rank : 195) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22561 | Gene names | Wt1, Wt-1 | |||
|
Domain Architecture |
|
|||||
| Description | Wilms' tumor protein homolog. | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.012367 (rank : 140) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
BBC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.020202 (rank : 123) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXH1, O00171, Q96PG9 | Gene names | BBC3, PUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-binding component 3 (p53 up-regulated modulator of apoptosis) (JFY-1). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.013922 (rank : 134) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.016609 (rank : 132) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.009220 (rank : 149) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
K0552_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.018295 (rank : 128) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.020374 (rank : 122) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.063901 (rank : 53) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.146078 (rank : 33) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.028204 (rank : 107) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
WT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | -0.000704 (rank : 196) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19544, Q15881, Q16256, Q16575, Q8IYZ5 | Gene names | WT1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein (WT33). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.005798 (rank : 171) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.024253 (rank : 116) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
CNOT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.018783 (rank : 126) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN8, Q9H3E0, Q9NSX5, Q9NWR6, Q9P028 | Gene names | CNOT2, CDC36, NOT2 | |||
|
Domain Architecture |
|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
CNOT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.018609 (rank : 127) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5L3, Q3UE39, Q80YA5, Q9D0P1 | Gene names | Cnot2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.008731 (rank : 150) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
DC1L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.009234 (rank : 148) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1Q8 | Gene names | Dync1li1, Dncli1, Dnclic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
FBX10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.014885 (rank : 133) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK96, Q5JRT8, Q9UKC3 | Gene names | FBXO10, FBX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 10. | |||||
|
HTF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.012378 (rank : 139) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
ISL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.003634 (rank : 181) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61371, P20663, P47894 | Gene names | ISL1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.003634 (rank : 182) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61372, P20663, P47894, Q812D8 | Gene names | Isl1 | |||
|
Domain Architecture |
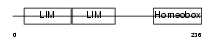
|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
P73L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.020743 (rank : 121) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.138942 (rank : 35) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.030130 (rank : 104) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.024392 (rank : 115) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ST1C1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.004589 (rank : 179) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VR3, O70262 | Gene names | Sult1c1, Sult1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 1C1 (EC 2.8.2.-) (Phenol sulfotransferase). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.012285 (rank : 141) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.011913 (rank : 143) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
APOA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.009879 (rank : 146) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6Q788, Q6UWK9, Q9UBJ3 | Gene names | APOA5, RAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apolipoprotein A-V precursor (Apolipoprotein A5) (ApoAV) (ApoA-V) (Regeneration-associated protein 3). | |||||
|
CSTF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.006456 (rank : 167) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |
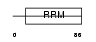
|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
EAN57_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.012431 (rank : 138) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.023091 (rank : 118) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
KAISO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | -0.000289 (rank : 194) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BN78, Q8C226, Q9WTZ7 | Gene names | Zbtb33, Kaiso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
MDR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.006111 (rank : 169) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21447 | Gene names | Abcb1a, Abcb4, Mdr1a, Mdr3, Pgy-3, Pgy3 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 3 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1A) (P-glycoprotein 3) (MDR1A). | |||||
|
NKX61_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.002787 (rank : 188) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.007627 (rank : 159) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.025806 (rank : 111) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.056895 (rank : 60) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T3JAM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.006367 (rank : 168) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.037302 (rank : 93) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.020968 (rank : 120) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
ZBT46_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.000761 (rank : 192) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UZ6, Q5JWJ3, Q9BQK3, Q9H3Z8, Q9H3Z9 | Gene names | ZBTB46, BTBD4, ZNF340 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 46 (BTB/POZ domain- containing protein 4) (Zinc finger protein 340). | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.005849 (rank : 170) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ANDR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.008034 (rank : 154) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
ARHG9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.005275 (rank : 176) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
ARHG9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.005287 (rank : 174) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.003561 (rank : 184) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.003387 (rank : 185) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.001785 (rank : 190) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
BTBD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.002989 (rank : 187) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C726 | Gene names | Btbd9 | |||
|
Domain Architecture |
|
|||||
| Description | BTB/POZ domain-containing protein 9. | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.027233 (rank : 110) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.007541 (rank : 160) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
FXR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.007704 (rank : 157) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
GO45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.007754 (rank : 156) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2X8, Q3TJC4, Q8C0U6, Q9DAV7 | Gene names | Blzf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin 45 (Basic leucine zipper nuclear factor 1). | |||||
|
HES7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.051778 (rank : 76) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.049249 (rank : 82) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.078930 (rank : 42) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.002049 (rank : 189) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MDR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.005279 (rank : 175) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06795 | Gene names | Abcb1, Abcb1b, Mdr1, Mdr1b, Pgy1, Pgy1-1 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 1 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1) (P-glycoprotein 1) (CD243 antigen). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.042138 (rank : 89) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.009902 (rank : 145) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.027466 (rank : 108) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.007627 (rank : 158) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.027392 (rank : 109) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.051103 (rank : 77) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.052845 (rank : 70) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.053707 (rank : 69) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.064489 (rank : 51) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.067458 (rank : 49) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.072306 (rank : 44) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.070662 (rank : 45) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.066361 (rank : 50) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.067541 (rank : 48) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.060993 (rank : 54) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060957 (rank : 55) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
PER1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.143416 (rank : 34) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.131575 (rank : 37) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.132238 (rank : 36) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.122852 (rank : 38) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.056578 (rank : 61) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.052364 (rank : 72) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.059109 (rank : 57) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.056073 (rank : 63) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.052152 (rank : 75) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.054129 (rank : 68) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.052237 (rank : 73) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.052212 (rank : 74) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.050448 (rank : 80) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.050453 (rank : 79) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 171 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.966968 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.934324 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.932488 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.912004 (rank : 5) | θ value | 2.36115e-125 (rank : 6) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.910960 (rank : 6) | θ value | 3.5201e-129 (rank : 5) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.605117 (rank : 7) | θ value | 9.90251e-15 (rank : 11) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.604957 (rank : 8) | θ value | 2.5182e-18 (rank : 8) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.603881 (rank : 9) | θ value | 5.07402e-19 (rank : 7) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.602801 (rank : 10) | θ value | 1.68911e-14 (rank : 12) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.589777 (rank : 11) | θ value | 3.52202e-12 (rank : 16) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.582022 (rank : 12) | θ value | 2.0648e-12 (rank : 15) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.575044 (rank : 13) | θ value | 4.44505e-15 (rank : 9) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.574792 (rank : 14) | θ value | 5.8054e-15 (rank : 10) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.572833 (rank : 15) | θ value | 2.13673e-09 (rank : 24) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.569862 (rank : 16) | θ value | 7.84624e-12 (rank : 17) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.569420 (rank : 17) | θ value | 3.64472e-09 (rank : 25) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.566854 (rank : 18) | θ value | 6.64225e-11 (rank : 19) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.566111 (rank : 19) | θ value | 2.28291e-11 (rank : 18) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.565466 (rank : 20) | θ value | 1.47974e-10 (rank : 20) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.563631 (rank : 21) | θ value | 6.41864e-14 (rank : 13) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.562303 (rank : 22) | θ value | 4.30538e-10 (rank : 22) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.561427 (rank : 23) | θ value | 4.30538e-10 (rank : 23) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.555783 (rank : 24) | θ value | 3.29651e-10 (rank : 21) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.555782 (rank : 25) | θ value | 4.16044e-13 (rank : 14) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.553234 (rank : 26) | θ value | 1.38499e-08 (rank : 26) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.537310 (rank : 27) | θ value | 6.87365e-08 (rank : 27) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.527169 (rank : 28) | θ value | 2.61198e-07 (rank : 28) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.524212 (rank : 29) | θ value | 7.59969e-07 (rank : 30) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.521640 (rank : 30) | θ value | 7.59969e-07 (rank : 29) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.364593 (rank : 31) | θ value | 0.0252991 (rank : 38) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.357377 (rank : 32) | θ value | 0.0252991 (rank : 39) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.146078 (rank : 33) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.143416 (rank : 34) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.138942 (rank : 35) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.132238 (rank : 36) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.131575 (rank : 37) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.122852 (rank : 38) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.117686 (rank : 39) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.115727 (rank : 40) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.084330 (rank : 41) | θ value | 0.0330416 (rank : 41) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.078930 (rank : 42) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.074741 (rank : 43) | θ value | 0.00390308 (rank : 31) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.072306 (rank : 44) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.070662 (rank : 45) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
SSXT_HUMAN
|
||||||
| NC score | 0.069481 (rank : 46) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
SSXT_MOUSE
|
||||||
| NC score | 0.067858 (rank : 47) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.067541 (rank : 48) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.067458 (rank : 49) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.066361 (rank : 50) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.064489 (rank : 51) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.064119 (rank : 52) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.063901 (rank : 53) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.060993 (rank : 54) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.060957 (rank : 55) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
TEX15_HUMAN
|
||||||
| NC score | 0.059380 (rank : 56) | θ value | 0.0193708 (rank : 36) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.059109 (rank : 57) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.057454 (rank : 58) | θ value | 0.0193708 (rank : 34) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.057454 (rank : 59) | θ value | 0.0193708 (rank : 35) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.056895 (rank : 60) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.056578 (rank : 61) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.056318 (rank : 62) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.056073 (rank : 63) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.055896 (rank : 64) | θ value | 0.0148317 (rank : 33) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.055713 (rank : 65) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.055581 (rank : 66) | θ value | 0.0252991 (rank : 37) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.055532 (rank : 67) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.054129 (rank : 68) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.053707 (rank : 69) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.052845 (rank : 70) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MAML1_HUMAN
|
||||||
| NC score | 0.052500 (rank : 71) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.052364 (rank : 72) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.052237 (rank : 73) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.052212 (rank : 74) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.052152 (rank : 75) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.051778 (rank : 76) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.051103 (rank : 77) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.051073 (rank : 78) | θ value | 0.00869519 (rank : 32) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.050453 (rank : 79) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.050448 (rank : 80) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.049435 (rank : 81) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.049249 (rank : 82) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.049176 (rank : 83) | θ value | 0.163984 (rank : 48) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.047391 (rank : 84) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.047161 (rank : 85) | θ value | 0.47712 (rank : 65) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.047011 (rank : 86) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.046753 (rank : 87) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.045412 (rank : 88) | θ value | 0.279714 (rank : 57) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.042138 (rank : 89) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.041869 (rank : 90) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.039918 (rank : 91) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.037597 (rank : 92) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.037302 (rank : 93) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.036146 (rank : 94) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.035920 (rank : 95) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.035474 (rank : 96) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
N4BP2_HUMAN
|
||||||
| NC score | 0.033380 (rank : 97) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.032960 (rank : 98) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.032768 (rank : 99) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.032019 (rank : 100) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.031964 (rank : 101) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.031350 (rank : 102) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.030909 (rank : 103) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.030130 (rank : 104) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
K0240_HUMAN
|
||||||
| NC score | 0.029246 (rank : 105) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.028485 (rank : 106) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.028204 (rank : 107) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.027466 (rank : 108) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.027392 (rank : 109) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.027233 (rank : 110) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.025806 (rank : 111) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.025280 (rank : 112) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
SNX15_MOUSE
|
||||||
| NC score | 0.024819 (rank : 113) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.024470 (rank : 114) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.024392 (rank : 115) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.024253 (rank : 116) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.024046 (rank : 117) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.023091 (rank : 118) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.022716 (rank : 119) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.020968 (rank : 120) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.020743 (rank : 121) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.020374 (rank : 122) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
BBC3_HUMAN
|
||||||
| NC score | 0.020202 (rank : 123) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXH1, O00171, Q96PG9 | Gene names | BBC3, PUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-binding component 3 (p53 up-regulated modulator of apoptosis) (JFY-1). | |||||
|
USBP1_MOUSE
|
||||||
| NC score | 0.019767 (rank : 124) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.019265 (rank : 125) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
CNOT2_HUMAN
|
||||||
| NC score | 0.018783 (rank : 126) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN8, Q9H3E0, Q9NSX5, Q9NWR6, Q9P028 | Gene names | CNOT2, CDC36, NOT2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
CNOT2_MOUSE
|
||||||
| NC score | 0.018609 (rank : 127) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5L3, Q3UE39, Q80YA5, Q9D0P1 | Gene names | Cnot2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
K0552_HUMAN
|
||||||
| NC score | 0.018295 (rank : 128) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.017641 (rank : 129) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.017404 (rank : 130) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.017117 (rank : 131) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.016609 (rank : 132) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
FBX10_HUMAN
|
||||||
| NC score | 0.014885 (rank : 133) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK96, Q5JRT8, Q9UKC3 | Gene names | FBXO10, FBX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 10. | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.013922 (rank : 134) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.013659 (rank : 135) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
INP5E_MOUSE
|
||||||
| NC score | 0.013545 (rank : 136) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
TTLL4_MOUSE
|
||||||
| NC score | 0.012444 (rank : 137) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
EAN57_HUMAN
|
||||||
| NC score | 0.012431 (rank : 138) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
HTF4_MOUSE
|
||||||
| NC score | 0.012378 (rank : 139) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.012367 (rank : 140) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.012285 (rank : 141) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.012038 (rank : 142) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.011913 (rank : 143) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.010482 (rank : 144) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.009902 (rank : 145) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
APOA5_HUMAN
|
||||||
| NC score | 0.009879 (rank : 146) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6Q788, Q6UWK9, Q9UBJ3 | Gene names | APOA5, RAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apolipoprotein A-V precursor (Apolipoprotein A5) (ApoAV) (ApoA-V) (Regeneration-associated protein 3). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.009446 (rank : 147) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
DC1L1_MOUSE
|
||||||
| NC score | 0.009234 (rank : 148) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1Q8 | Gene names | Dync1li1, Dncli1, Dnclic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.009220 (rank : 149) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.008731 (rank : 150) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.008686 (rank : 151) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
SUHW4_MOUSE
|
||||||
| NC score | 0.008510 (rank : 152) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.008148 (rank : 153) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
ANDR_MOUSE
|
||||||
| NC score | 0.008034 (rank : 154) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.007935 (rank : 155) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
GO45_MOUSE
|
||||||
| NC score | 0.007754 (rank : 156) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2X8, Q3TJC4, Q8C0U6, Q9DAV7 | Gene names | Blzf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin 45 (Basic leucine zipper nuclear factor 1). | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.007704 (rank : 157) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.007627 (rank : 158) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.007627 (rank : 159) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.007541 (rank : 160) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.007541 (rank : 161) | θ value | 0.0252991 (rank : 40) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ABCBB_HUMAN
|
||||||
| NC score | 0.007416 (rank : 162) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95342, Q9UNB2 | Gene names | ABCB11, BSEP | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt export pump (ATP-binding cassette sub-family B member 11). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.007137 (rank : 163) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.007060 (rank : 164) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.006732 (rank : 165) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.006530 (rank : 166) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CSTF2_HUMAN
|
||||||
| NC score | 0.006456 (rank : 167) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
T3JAM_HUMAN
|
||||||
| NC score | 0.006367 (rank : 168) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
MDR3_MOUSE
|
||||||
| NC score | 0.006111 (rank : 169) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21447 | Gene names | Abcb1a, Abcb4, Mdr1a, Mdr3, Pgy-3, Pgy3 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 3 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1A) (P-glycoprotein 3) (MDR1A). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.005849 (rank : 170) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.005798 (rank : 171) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.005469 (rank : 172) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
TAOK1_MOUSE
|
||||||
| NC score | 0.005367 (rank : 173) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
ARHG9_MOUSE
|
||||||
| NC score | 0.005287 (rank : 174) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
MDR1_MOUSE
|
||||||
| NC score | 0.005279 (rank : 175) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06795 | Gene names | Abcb1, Abcb1b, Mdr1, Mdr1b, Pgy1, Pgy1-1 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 1 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1) (P-glycoprotein 1) (CD243 antigen). | |||||
|
ARHG9_HUMAN
|
||||||
| NC score | 0.005275 (rank : 176) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
PCDA7_HUMAN
|
||||||
| NC score | 0.005096 (rank : 177) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
TAOK1_HUMAN
|
||||||
| NC score | 0.005056 (rank : 178) | θ value | 0.47712 (rank : 66) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
ST1C1_MOUSE
|
||||||
| NC score | 0.004589 (rank : 179) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VR3, O70262 | Gene names | Sult1c1, Sult1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 1C1 (EC 2.8.2.-) (Phenol sulfotransferase). | |||||
|
ZBT46_MOUSE
|
||||||
| NC score | 0.004004 (rank : 180) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BID6, Q3U2R2, Q9D0G6, Q9D492 | Gene names | Zbtb46, Btbd4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 46 (BTB/POZ domain- containing protein 4). | |||||
|
ISL1_HUMAN
|
||||||
| NC score | 0.003634 (rank : 181) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61371, P20663, P47894 | Gene names | ISL1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL1_MOUSE
|
||||||
| NC score | 0.003634 (rank : 182) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61372, P20663, P47894, Q812D8 | Gene names | Isl1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
PCDA5_HUMAN
|
||||||
| NC score | 0.003587 (rank : 183) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.003561 (rank : 184) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| NC score | 0.003387 (rank : 185) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.003151 (rank : 186) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
BTBD9_MOUSE
|
||||||
| NC score | 0.002989 (rank : 187) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C726 | Gene names | Btbd9 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 9. | |||||
|
NKX61_MOUSE
|
||||||
| NC score | 0.002787 (rank : 188) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.002049 (rank : 189) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.001785 (rank : 190) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
PRD16_HUMAN
|
||||||
| NC score | 0.000772 (rank : 191) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
ZBT46_HUMAN
|
||||||
| NC score | 0.000761 (rank : 192) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UZ6, Q5JWJ3, Q9BQK3, Q9H3Z8, Q9H3Z9 | Gene names | ZBTB46, BTBD4, ZNF340 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 46 (BTB/POZ domain- containing protein 4) (Zinc finger protein 340). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.000199 (rank : 193) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
KAISO_MOUSE
|
||||||
| NC score | -0.000289 (rank : 194) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BN78, Q8C226, Q9WTZ7 | Gene names | Zbtb33, Kaiso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
WT1_MOUSE
|
||||||
| NC score | -0.000537 (rank : 195) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22561 | Gene names | Wt1, Wt-1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein homolog. | |||||
|
WT1_HUMAN
|
||||||
| NC score | -0.000704 (rank : 196) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19544, Q15881, Q16256, Q16575, Q8IYZ5 | Gene names | WT1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein (WT33). | |||||