Please be patient as the page loads
|
GGA2_HUMAN
|
||||||
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GGA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988972 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 1.38103e-133 (rank : 3) | NC score | 0.948066 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 3.4016e-132 (rank : 4) | NC score | 0.949589 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 1.95407e-87 (rank : 5) | NC score | 0.932343 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 1.65422e-86 (rank : 6) | NC score | 0.920737 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 7) | NC score | 0.651067 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 8) | NC score | 0.666493 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 9) | NC score | 0.637563 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TOM1_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 10) | NC score | 0.661576 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 11) | NC score | 0.382143 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 12) | NC score | 0.389847 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 13) | NC score | 0.375134 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 14) | NC score | 0.365670 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 15) | NC score | 0.390753 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 16) | NC score | 0.392699 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 17) | NC score | 0.392029 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 18) | NC score | 0.375123 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 19) | NC score | 0.380185 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 20) | NC score | 0.375251 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 21) | NC score | 0.011643 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.020665 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.024834 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.049361 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.058059 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.044220 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.045333 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.033964 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.018916 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.035282 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CO027_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.037011 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BZB3, Q6NVG3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27 homolog. | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.018627 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
ESPL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.026771 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.012617 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
K0240_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.041673 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.022634 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.020651 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.005559 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.032462 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.002444 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.032423 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NFIA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.021120 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12857, Q9H3X9, Q9P2A9 | Gene names | NFIA, KIAA1439 | |||
|
Domain Architecture |
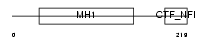
|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.031998 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.025161 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.017699 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
VGF_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.037092 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
BCDO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.024768 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAY6, Q9NVH5 | Gene names | BCMO1, BCDO, BCDO1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta,beta-carotene 15,15'-monooxygenase (EC 1.14.99.36) (Beta-carotene dioxygenase 1). | |||||
|
HNF1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.033116 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.018251 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.020363 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.020101 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.027953 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.006042 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BCDO1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.023828 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJS6, Q6K1L5, Q8C6N5, Q9ERN9 | Gene names | Bcmo1, Bcdo, Bcdo1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta,beta-carotene 15,15'-monooxygenase (EC 1.14.99.36) (Beta-carotene dioxygenase 1). | |||||
|
K0240_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.036701 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
K0555_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.021342 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.026363 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
ROBO3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.006298 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.013444 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
DAAM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.012798 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
ECM29_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.026744 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
NFIA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.019328 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02780, P70250, P70251, Q3UUZ2, Q61960, Q8VBT5, Q9R1G5 | Gene names | Nfia | |||
|
Domain Architecture |
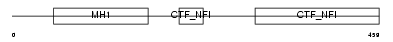
|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.021021 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
STAP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.020535 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.029337 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.017763 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.022930 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CN092_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.026313 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
ECM29_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.025545 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYK3, O15074, Q8WU82 | Gene names | ECM29, KIAA0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
IRX4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.018607 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QY61 | Gene names | Irx4, Irxa3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-4 (Iroquois homeobox protein 4) (Homeodomain protein IRXA3). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.016612 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC8C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.007571 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R502, Q3TEP9, Q8C296, Q8R0N7, Q8R3G5 | Gene names | Lrrc8c, Ad158 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8C (Protein AD158). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.012245 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.019469 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.018358 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.009861 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
HCLS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.034206 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
MRP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.006017 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI47 | Gene names | Abcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 1 (ATP-binding cassette sub-family C member 2). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.016609 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.002538 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
A4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.014742 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.015378 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.004286 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.013909 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.001463 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.028742 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.008100 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.010740 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.011349 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.028742 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.011572 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.009012 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.028752 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
INADL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.003796 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
MEF2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.007237 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |
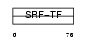
|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.011875 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.004529 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.014275 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
TIP60_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.011362 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
TIP60_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.011486 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.014023 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
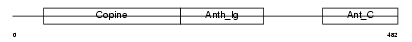
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.003104 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CRBB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.008317 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53674 | Gene names | CRYBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B1. | |||||
|
FOXO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.005331 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1E0 | Gene names | Foxo1a, Fkhr, Foxo1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.006861 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.030978 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RAVR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.005679 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
SLIK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.001109 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
SYMPK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.011719 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.009543 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TSSP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.008124 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQE7, O75416 | Gene names | PRSS16, TSSP | |||
|
Domain Architecture |

|
|||||
| Description | Thymus-specific serine protease precursor (EC 3.4.-.-). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.009815 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZN592_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | -0.001029 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
ZN692_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | -0.002151 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.106404 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.104267 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.104121 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.104360 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.091904 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.096689 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.108309 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.107974 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.988972 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.949589 (rank : 3) | θ value | 3.4016e-132 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.948066 (rank : 4) | θ value | 1.38103e-133 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.932343 (rank : 5) | θ value | 1.95407e-87 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.920737 (rank : 6) | θ value | 1.65422e-86 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.666493 (rank : 7) | θ value | 3.07829e-16 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.661576 (rank : 8) | θ value | 1.16975e-15 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.651067 (rank : 9) | θ value | 6.20254e-17 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.637563 (rank : 10) | θ value | 6.85773e-16 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.392699 (rank : 11) | θ value | 3.29651e-10 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.392029 (rank : 12) | θ value | 5.62301e-10 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.390753 (rank : 13) | θ value | 1.9326e-10 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.389847 (rank : 14) | θ value | 2.20605e-14 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.382143 (rank : 15) | θ value | 1.29331e-14 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.380185 (rank : 16) | θ value | 1.38499e-08 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.375251 (rank : 17) | θ value | 1.80886e-08 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.375134 (rank : 18) | θ value | 2.43908e-13 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.375123 (rank : 19) | θ value | 4.76016e-09 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.365670 (rank : 20) | θ value | 1.33837e-11 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.108309 (rank : 21) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.107974 (rank : 22) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.106404 (rank : 23) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.104360 (rank : 24) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.104267 (rank : 25) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.104121 (rank : 26) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.096689 (rank : 27) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.091904 (rank : 28) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.058059 (rank : 29) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.049361 (rank : 30) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.045333 (rank : 31) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.044220 (rank : 32) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
K0240_MOUSE
|
||||||
| NC score | 0.041673 (rank : 33) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
VGF_HUMAN
|
||||||
| NC score | 0.037092 (rank : 34) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
CO027_MOUSE
|
||||||
| NC score | 0.037011 (rank : 35) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BZB3, Q6NVG3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27 homolog. | |||||
|
K0240_HUMAN
|
||||||
| NC score | 0.036701 (rank : 36) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.035282 (rank : 37) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
HCLS1_HUMAN
|
||||||
| NC score | 0.034206 (rank : 38) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.033964 (rank : 39) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
HNF1A_HUMAN
|
||||||
| NC score | 0.033116 (rank : 40) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.032462 (rank : 41) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.032423 (rank : 42) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.031998 (rank : 43) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.030978 (rank : 44) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.029337 (rank : 45) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.028752 (rank : 46) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.028742 (rank : 47) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.028742 (rank : 48) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.027953 (rank : 49) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
ESPL1_MOUSE
|
||||||
| NC score | 0.026771 (rank : 50) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
ECM29_MOUSE
|
||||||
| NC score | 0.026744 (rank : 51) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.026363 (rank : 52) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.026313 (rank : 53) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
ECM29_HUMAN
|
||||||
| NC score | 0.025545 (rank : 54) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYK3, O15074, Q8WU82 | Gene names | ECM29, KIAA0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.025161 (rank : 55) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.024834 (rank : 56) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
BCDO1_HUMAN
|
||||||
| NC score | 0.024768 (rank : 57) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAY6, Q9NVH5 | Gene names | BCMO1, BCDO, BCDO1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta,beta-carotene 15,15'-monooxygenase (EC 1.14.99.36) (Beta-carotene dioxygenase 1). | |||||
|
BCDO1_MOUSE
|
||||||
| NC score | 0.023828 (rank : 58) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJS6, Q6K1L5, Q8C6N5, Q9ERN9 | Gene names | Bcmo1, Bcdo, Bcdo1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta,beta-carotene 15,15'-monooxygenase (EC 1.14.99.36) (Beta-carotene dioxygenase 1). | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.022930 (rank : 59) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.022634 (rank : 60) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.021342 (rank : 61) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
NFIA_HUMAN
|
||||||
| NC score | 0.021120 (rank : 62) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12857, Q9H3X9, Q9P2A9 | Gene names | NFIA, KIAA1439 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.021021 (rank : 63) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.020665 (rank : 64) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.020651 (rank : 65) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
STAP2_MOUSE
|
||||||
| NC score | 0.020535 (rank : 66) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.020363 (rank : 67) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.020101 (rank : 68) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.019469 (rank : 69) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
NFIA_MOUSE
|
||||||
| NC score | 0.019328 (rank : 70) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02780, P70250, P70251, Q3UUZ2, Q61960, Q8VBT5, Q9R1G5 | Gene names | Nfia | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.018916 (rank : 71) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.018627 (rank : 72) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
IRX4_MOUSE
|
||||||
| NC score | 0.018607 (rank : 73) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QY61 | Gene names | Irx4, Irxa3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-4 (Iroquois homeobox protein 4) (Homeodomain protein IRXA3). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.018358 (rank : 74) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.018251 (rank : 75) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.017763 (rank : 76) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.017699 (rank : 77) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.016612 (rank : 78) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.016609 (rank : 79) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.015378 (rank : 80) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.014742 (rank : 81) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.014275 (rank : 82) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.014023 (rank : 83) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.013909 (rank : 84) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.013444 (rank : 85) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
DAAM2_MOUSE
|
||||||
| NC score | 0.012798 (rank : 86) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.012617 (rank : 87) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.012245 (rank : 88) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.011875 (rank : 89) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SYMPK_HUMAN
|
||||||
| NC score | 0.011719 (rank : 90) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.011643 (rank : 91) | θ value | 0.0193708 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.011572 (rank : 92) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
TIP60_MOUSE
|
||||||
| NC score | 0.011486 (rank : 93) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
TIP60_HUMAN
|
||||||
| NC score | 0.011362 (rank : 94) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.011349 (rank : 95) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.010740 (rank : 96) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.009861 (rank : 97) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.009815 (rank : 98) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.009543 (rank : 99) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.009012 (rank : 100) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CRBB1_HUMAN
|
||||||
| NC score | 0.008317 (rank : 101) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53674 | Gene names | CRYBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B1. | |||||
|
TSSP_HUMAN
|
||||||
| NC score | 0.008124 (rank : 102) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQE7, O75416 | Gene names | PRSS16, TSSP | |||
|
Domain Architecture |

|
|||||
| Description | Thymus-specific serine protease precursor (EC 3.4.-.-). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.008100 (rank : 103) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
LRC8C_MOUSE
|
||||||
| NC score | 0.007571 (rank : 104) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R502, Q3TEP9, Q8C296, Q8R0N7, Q8R3G5 | Gene names | Lrrc8c, Ad158 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8C (Protein AD158). | |||||
|
MEF2A_MOUSE
|
||||||
| NC score | 0.007237 (rank : 105) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.006861 (rank : 106) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
ROBO3_MOUSE
|
||||||
| NC score | 0.006298 (rank : 107) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.006042 (rank : 108) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
MRP2_MOUSE
|
||||||
| NC score | 0.006017 (rank : 109) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI47 | Gene names | Abcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 1 (ATP-binding cassette sub-family C member 2). | |||||
|
RAVR1_MOUSE
|
||||||
| NC score | 0.005679 (rank : 110) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.005559 (rank : 111) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
FOXO1_MOUSE
|
||||||
| NC score | 0.005331 (rank : 112) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1E0 | Gene names | Foxo1a, Fkhr, Foxo1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.004529 (rank : 113) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.004286 (rank : 114) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.003796 (rank : 115) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.003104 (rank : 116) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.002538 (rank : 117) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.002444 (rank : 118) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.001463 (rank : 119) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
SLIK3_HUMAN
|
||||||
| NC score | 0.001109 (rank : 120) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
ZN592_MOUSE
|
||||||
| NC score | -0.001029 (rank : 121) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
ZN692_HUMAN
|
||||||
| NC score | -0.002151 (rank : 122) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||