Please be patient as the page loads
|
AP2A1_MOUSE
|
||||||
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AP2A1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996547 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.993685 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993175 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 3.34864e-71 (rank : 5) | NC score | 0.841814 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 3.34864e-71 (rank : 6) | NC score | 0.844862 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 1.45591e-66 (rank : 7) | NC score | 0.850590 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 4.23606e-66 (rank : 8) | NC score | 0.841455 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | 2.95404e-43 (rank : 9) | NC score | 0.808548 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 10) | NC score | 0.809083 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 11) | NC score | 0.761273 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 12) | NC score | 0.736858 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 13) | NC score | 0.473435 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 14) | NC score | 0.473449 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP1B1_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 15) | NC score | 0.476931 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 16) | NC score | 0.475375 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.033759 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.017332 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.005174 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.020389 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.030958 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.022735 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.032793 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.025371 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.012253 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.025044 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.016613 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.026519 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.014597 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.012521 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.012739 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.007071 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.019174 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.003849 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
AP4B1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.250690 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.015828 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.004742 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
EGR4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | -0.000987 (rank : 72) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
MGRN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.012795 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
CU005_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.016553 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3R5, Q9UEZ3 | Gene names | C21orf5, KIAA0933 | |||
|
Domain Architecture |
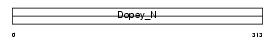
|
|||||
| Description | Protein C21orf5. | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.012361 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.004154 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ANR25_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.008077 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.009933 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CFAD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.000871 (rank : 71) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00746 | Gene names | CFD, DF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin). | |||||
|
NEK11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.002372 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.011565 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.011747 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SSH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.006664 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
STAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.007800 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.024246 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
AP4B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.229519 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.009992 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.011305 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
RIB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.012073 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YQ5, Q3U985 | Gene names | Rpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I). | |||||
|
SYYC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.016686 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54577, O43276, Q53EN1 | Gene names | YARS | |||
|
Domain Architecture |
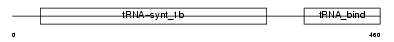
|
|||||
| Description | Tyrosyl-tRNA synthetase, cytoplasmic (EC 6.1.1.1) (Tyrosyl--tRNA ligase) (TyrRS). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.003087 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.195956 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.195534 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.162869 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.163410 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
COPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.220058 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.220548 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.114828 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.122486 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.109712 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.091228 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.089195 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.104267 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.108116 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.100293 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.102301 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.996547 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.993685 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.993175 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.850590 (rank : 5) | θ value | 1.45591e-66 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.844862 (rank : 6) | θ value | 3.34864e-71 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.841814 (rank : 7) | θ value | 3.34864e-71 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.841455 (rank : 8) | θ value | 4.23606e-66 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.809083 (rank : 9) | θ value | 1.46607e-42 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.808548 (rank : 10) | θ value | 2.95404e-43 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.761273 (rank : 11) | θ value | 1.79631e-32 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.736858 (rank : 12) | θ value | 3.06405e-32 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP1B1_MOUSE
|
||||||
| NC score | 0.476931 (rank : 13) | θ value | 1.80466e-16 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_HUMAN
|
||||||
| NC score | 0.475375 (rank : 14) | θ value | 3.07829e-16 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP2B1_MOUSE
|
||||||
| NC score | 0.473449 (rank : 15) | θ value | 6.20254e-17 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.473435 (rank : 16) | θ value | 6.20254e-17 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP4B1_MOUSE
|
||||||
| NC score | 0.250690 (rank : 17) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP4B1_HUMAN
|
||||||
| NC score | 0.229519 (rank : 18) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
COPB_MOUSE
|
||||||
| NC score | 0.220548 (rank : 19) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_HUMAN
|
||||||
| NC score | 0.220058 (rank : 20) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.195956 (rank : 21) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.195534 (rank : 22) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.163410 (rank : 23) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.162869 (rank : 24) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
COPG2_MOUSE
|
||||||
| NC score | 0.122486 (rank : 25) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG2_HUMAN
|
||||||
| NC score | 0.114828 (rank : 26) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| NC score | 0.109712 (rank : 27) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.108116 (rank : 28) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.104267 (rank : 29) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.102301 (rank : 30) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.100293 (rank : 31) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.091228 (rank : 32) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.089195 (rank : 33) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.033759 (rank : 34) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.032793 (rank : 35) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.030958 (rank : 36) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.026519 (rank : 37) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.025371 (rank : 38) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.025044 (rank : 39) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.024246 (rank : 40) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.022735 (rank : 41) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.020389 (rank : 42) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.019174 (rank : 43) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.017332 (rank : 44) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SYYC_HUMAN
|
||||||
| NC score | 0.016686 (rank : 45) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54577, O43276, Q53EN1 | Gene names | YARS | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-tRNA synthetase, cytoplasmic (EC 6.1.1.1) (Tyrosyl--tRNA ligase) (TyrRS). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.016613 (rank : 46) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CU005_HUMAN
|
||||||
| NC score | 0.016553 (rank : 47) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3R5, Q9UEZ3 | Gene names | C21orf5, KIAA0933 | |||
|
Domain Architecture |
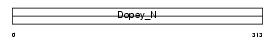
|
|||||
| Description | Protein C21orf5. | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.015828 (rank : 48) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.014597 (rank : 49) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
MGRN1_HUMAN
|
||||||
| NC score | 0.012795 (rank : 50) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.012739 (rank : 51) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.012521 (rank : 52) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.012361 (rank : 53) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.012253 (rank : 54) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
RIB1_MOUSE
|
||||||
| NC score | 0.012073 (rank : 55) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YQ5, Q3U985 | Gene names | Rpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.011747 (rank : 56) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.011565 (rank : 57) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.011305 (rank : 58) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.009992 (rank : 59) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.009933 (rank : 60) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.008077 (rank : 61) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
STAT2_HUMAN
|
||||||
| NC score | 0.007800 (rank : 62) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.007071 (rank : 63) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.006664 (rank : 64) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.005174 (rank : 65) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.004742 (rank : 66) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.004154 (rank : 67) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.003849 (rank : 68) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.003087 (rank : 69) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
NEK11_HUMAN
|
||||||
| NC score | 0.002372 (rank : 70) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
CFAD_HUMAN
|
||||||
| NC score | 0.000871 (rank : 71) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00746 | Gene names | CFD, DF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin). | |||||
|
EGR4_MOUSE
|
||||||
| NC score | -0.000987 (rank : 72) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||