Please be patient as the page loads
|
GGA1_HUMAN
|
||||||
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GGA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.975245 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 3.4016e-132 (rank : 3) | NC score | 0.949589 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 2.06847e-121 (rank : 4) | NC score | 0.952646 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 1.29861e-115 (rank : 5) | NC score | 0.931614 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 2.44908e-114 (rank : 6) | NC score | 0.943656 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 7) | NC score | 0.687548 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 8) | NC score | 0.682721 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 9) | NC score | 0.652311 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 10) | NC score | 0.367836 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
TM1L1_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 11) | NC score | 0.639318 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 12) | NC score | 0.358548 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 13) | NC score | 0.405825 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 14) | NC score | 0.404895 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 15) | NC score | 0.384835 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 16) | NC score | 0.389598 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 17) | NC score | 0.348564 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 18) | NC score | 0.395639 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 19) | NC score | 0.381041 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 20) | NC score | 0.339757 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 21) | NC score | 0.040146 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 22) | NC score | 0.071891 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 23) | NC score | 0.071133 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 24) | NC score | 0.059119 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 25) | NC score | 0.059046 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 26) | NC score | 0.071231 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
DACH1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.038052 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.040766 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.030097 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.014909 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.015290 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.043702 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.058063 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.028950 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.017320 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.030323 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
COEA1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.022582 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.025881 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.038091 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.024638 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.029454 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.037123 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.036468 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.037009 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
BRSK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.003907 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
COEA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.021144 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.028820 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.014121 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
RTN4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.035920 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
SOCS7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.024074 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
ANGL5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.019486 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.043855 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.018253 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.032077 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.024797 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.037329 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.032682 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.022582 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
IF39_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.022803 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.018751 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.024841 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.026362 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.044539 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.029143 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.029376 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
HCLS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.040503 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
K1958_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.028132 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.044829 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.029828 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.047769 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.020157 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ZBTB3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.004359 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.022652 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.045465 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.025184 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.005033 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.034177 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
YTHD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.018413 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.003243 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.035626 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.027678 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.042719 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.026286 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.012068 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.038358 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.023260 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
BORG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.018440 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
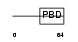
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
CASL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.022615 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |
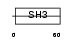
|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.020220 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.011215 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.004767 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
LIRB5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.006374 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.018305 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.029405 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.021331 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.020179 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.018304 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.009325 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
KRHB5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.008935 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78386, Q9NSB1 | Gene names | KRTHB5 | |||
|
Domain Architecture |
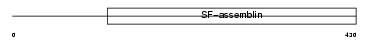
|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.009654 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MGRN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.012897 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D074, Q3U5V9, Q3UDA1, Q6ZQ97, Q8BZM9 | Gene names | Mgrn1, Kiaa0544 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.005073 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.008705 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.005124 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.021678 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.017437 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.016937 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
TM16H_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.014390 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
WEE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.003873 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30291 | Gene names | WEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase) (WEE1hu). | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.014512 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.006054 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ELL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.012700 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.014127 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.016822 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.024227 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.004575 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.004713 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TGIF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.008438 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZN2 | Gene names | TGIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.004557 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | -0.000516 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ZIC5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | -0.001516 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96T25 | Gene names | ZIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 5 (Zinc finger protein of the cerebellum 5). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.017626 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.011476 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
EXOC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.019412 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.016401 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FOXL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.002734 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12952, Q9H242 | Gene names | FOXL1, FKHL11, FREAC7 | |||
|
Domain Architecture |
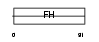
|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead- related transcription factor 7) (FREAC-7). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.032439 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
K1210_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.020023 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.002885 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.007667 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
NIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.019724 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
SC6A5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.002587 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y345, O95288, Q9BX77 | Gene names | SLC6A5, GLYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.016409 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.024659 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
WASP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.023023 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.018246 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.012659 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
ATF6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.011000 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.033181 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.034741 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.032256 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.027302 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.008447 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
DEN1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.015001 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.019979 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.009268 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EMIL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.009193 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99K41 | Gene names | Emilin1 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.016672 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
GLE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.018296 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.010233 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.003552 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.029524 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.008066 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
KREM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.004784 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.015911 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.007552 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.010630 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.013360 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.029249 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
SC24B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.010970 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.013993 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SF01_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.024096 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.006740 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.017130 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.031100 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.005370 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.009600 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.019445 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
ZYX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.010997 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.093760 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.091228 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.091001 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.092474 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.081867 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.085778 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.095177 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.095262 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.975245 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.952646 (rank : 3) | θ value | 2.06847e-121 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.949589 (rank : 4) | θ value | 3.4016e-132 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.943656 (rank : 5) | θ value | 2.44908e-114 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.931614 (rank : 6) | θ value | 1.29861e-115 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.687548 (rank : 7) | θ value | 7.0802e-21 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.682721 (rank : 8) | θ value | 1.2077e-20 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.652311 (rank : 9) | θ value | 1.16975e-15 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.639318 (rank : 10) | θ value | 6.41864e-14 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.405825 (rank : 11) | θ value | 1.02475e-11 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.404895 (rank : 12) | θ value | 1.74796e-11 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.395639 (rank : 13) | θ value | 3.64472e-09 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.389598 (rank : 14) | θ value | 3.29651e-10 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.384835 (rank : 15) | θ value | 1.9326e-10 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.381041 (rank : 16) | θ value | 6.21693e-09 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.367836 (rank : 17) | θ value | 1.29331e-14 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.358548 (rank : 18) | θ value | 3.52202e-12 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.348564 (rank : 19) | θ value | 5.62301e-10 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.339757 (rank : 20) | θ value | 1.06045e-08 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.095262 (rank : 21) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.095177 (rank : 22) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.093760 (rank : 23) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.092474 (rank : 24) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.091228 (rank : 25) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.091001 (rank : 26) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.085778 (rank : 27) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.081867 (rank : 28) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.071891 (rank : 29) | θ value | 0.00869519 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.071231 (rank : 30) | θ value | 0.0252991 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.071133 (rank : 31) | θ value | 0.0113563 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.059119 (rank : 32) | θ value | 0.0148317 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.059046 (rank : 33) | θ value | 0.0252991 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.058063 (rank : 34) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.047769 (rank : 35) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.045465 (rank : 36) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.044829 (rank : 37) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.044539 (rank : 38) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.043855 (rank : 39) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.043702 (rank : 40) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.042719 (rank : 41) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.040766 (rank : 42) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
HCLS1_MOUSE
|
||||||
| NC score | 0.040503 (rank : 43) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.040146 (rank : 44) | θ value | 0.00509761 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.038358 (rank : 45) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.038091 (rank : 46) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
DACH1_MOUSE
|
||||||
| NC score | 0.038052 (rank : 47) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.037329 (rank : 48) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.037123 (rank : 49) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.037009 (rank : 50) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.036468 (rank : 51) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
RTN4_HUMAN
|
||||||
| NC score | 0.035920 (rank : 52) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.035626 (rank : 53) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.034741 (rank : 54) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.034177 (rank : 55) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.033181 (rank : 56) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.032682 (rank : 57) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.032439 (rank : 58) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.032256 (rank : 59) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.032077 (rank : 60) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.031100 (rank : 61) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.030323 (rank : 62) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.030097 (rank : 63) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.029828 (rank : 64) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.029524 (rank : 65) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.029454 (rank : 66) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.029405 (rank : 67) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.029376 (rank : 68) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.029249 (rank : 69) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.029143 (rank : 70) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.028950 (rank : 71) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.028820 (rank : 72) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
K1958_HUMAN
|
||||||
| NC score | 0.028132 (rank : 73) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.027678 (rank : 74) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.027302 (rank : 75) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.026362 (rank : 76) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.026286 (rank : 77) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.025881 (rank : 78) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.025184 (rank : 79) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.024841 (rank : 80) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.024797 (rank : 81) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.024659 (rank : 82) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.024638 (rank : 83) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.024227 (rank : 84) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.024096 (rank : 85) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SOCS7_MOUSE
|
||||||
| NC score | 0.024074 (rank : 86) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.023260 (rank : 87) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.023023 (rank : 88) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.022803 (rank : 89) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.022652 (rank : 90) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
CASL_MOUSE
|
||||||
| NC score | 0.022615 (rank : 91) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.022582 (rank : 92) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.022582 (rank : 93) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.021678 (rank : 94) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.021331 (rank : 95) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
COEA1_HUMAN
|
||||||
| NC score | 0.021144 (rank : 96) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.020220 (rank : 97) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.020179 (rank : 98) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.020157 (rank : 99) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.020023 (rank : 100) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.019979 (rank : 101) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.019724 (rank : 102) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
ANGL5_HUMAN
|
||||||
| NC score | 0.019486 (rank : 103) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.019445 (rank : 104) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
EXOC1_HUMAN
|
||||||
| NC score | 0.019412 (rank : 105) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.018751 (rank : 106) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.018440 (rank : 107) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
YTHD1_HUMAN
|
||||||
| NC score | 0.018413 (rank : 108) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.018305 (rank : 109) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.018304 (rank : 110) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GLE1_MOUSE
|
||||||
| NC score | 0.018296 (rank : 111) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.018253 (rank : 112) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.018246 (rank : 113) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.017626 (rank : 114) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.017437 (rank : 115) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.017320 (rank : 116) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.017130 (rank : 117) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.016937 (rank : 118) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.016822 (rank : 119) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.016672 (rank : 120) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.016409 (rank : 121) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.016401 (rank : 122) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.015911 (rank : 123) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.015290 (rank : 124) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.015001 (rank : 125) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.014909 (rank : 126) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.014512 (rank : 127) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.014390 (rank : 128) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.014127 (rank : 129) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.014121 (rank : 130) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.013993 (rank : 131) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.013360 (rank : 132) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
MGRN1_MOUSE
|
||||||
| NC score | 0.012897 (rank : 133) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D074, Q3U5V9, Q3UDA1, Q6ZQ97, Q8BZM9 | Gene names | Mgrn1, Kiaa0544 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1). | |||||
|
ELL2_HUMAN
|
||||||
| NC score | 0.012700 (rank : 134) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.012659 (rank : 135) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.012068 (rank : 136) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.011476 (rank : 137) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.011215 (rank : 138) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
ATF6B_HUMAN
|
||||||
| NC score | 0.011000 (rank : 139) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
ZYX_HUMAN
|
||||||
| NC score | 0.010997 (rank : 140) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
SC24B_HUMAN
|
||||||
| NC score | 0.010970 (rank : 141) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.010630 (rank : 142) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.010233 (rank : 143) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.009654 (rank : 144) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.009600 (rank : 145) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.009325 (rank : 146) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.009268 (rank : 147) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EMIL1_MOUSE
|
||||||
| NC score | 0.009193 (rank : 148) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99K41 | Gene names | Emilin1 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
KRHB5_HUMAN
|
||||||
| NC score | 0.008935 (rank : 149) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78386, Q9NSB1 | Gene names | KRTHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.008705 (rank : 150) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.008447 (rank : 151) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
TGIF2_HUMAN
|
||||||
| NC score | 0.008438 (rank : 152) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZN2 | Gene names | TGIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.008066 (rank : 153) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.007667 (rank : 154) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.007552 (rank : 155) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.006740 (rank : 156) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
LIRB5_HUMAN
|
||||||
| NC score | 0.006374 (rank : 157) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.006054 (rank : 158) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.005370 (rank : 159) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.005124 (rank : 160) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.005073 (rank : 161) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.005033 (rank : 162) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
KREM1_MOUSE
|
||||||
| NC score | 0.004784 (rank : 163) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.004767 (rank : 164) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.004713 (rank : 165) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.004575 (rank : 166) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.004557 (rank : 167) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ZBTB3_HUMAN
|
||||||
| NC score | 0.004359 (rank : 168) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
BRSK2_HUMAN
|
||||||
| NC score | 0.003907 (rank : 169) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
WEE1_HUMAN
|
||||||
| NC score | 0.003873 (rank : 170) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30291 | Gene names | WEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase) (WEE1hu). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.003552 (rank : 171) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.003243 (rank : 172) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.002885 (rank : 173) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
FOXL1_HUMAN
|
||||||
| NC score | 0.002734 (rank : 174) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12952, Q9H242 | Gene names | FOXL1, FKHL11, FREAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead- related transcription factor 7) (FREAC-7). | |||||
|
SC6A5_HUMAN
|
||||||
| NC score | 0.002587 (rank : 175) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y345, O95288, Q9BX77 | Gene names | SLC6A5, GLYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.000516 (rank : 176) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ZIC5_HUMAN
|
||||||
| NC score | -0.001516 (rank : 177) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96T25 | Gene names | ZIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 5 (Zinc finger protein of the cerebellum 5). | |||||