Please be patient as the page loads
|
TM16H_HUMAN
|
||||||
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TM16H_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
TM16H_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.953761 (rank : 2) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
TM16E_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 3) | NC score | 0.689390 (rank : 3) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q75UR0, Q8CC04 | Gene names | Tmem16e, Gdd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16E (Gnathodiaphyseal dysplasia 1 protein homolog). | |||||
|
TM16F_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 4) | NC score | 0.686335 (rank : 4) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P9J9, Q8C242 | Gene names | Tmem16f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16F. | |||||
|
TM16E_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 5) | NC score | 0.683896 (rank : 7) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q75V66 | Gene names | TMEM16E, GDD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16E (Gnathodiaphyseal dysplasia 1 protein). | |||||
|
TM16F_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 6) | NC score | 0.684819 (rank : 5) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q4KMQ2, Q8N3Q2 | Gene names | TMEM16F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16F. | |||||
|
TM16C_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 7) | NC score | 0.683996 (rank : 6) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYT9 | Gene names | TMEM16C, C11orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protein 16C. | |||||
|
TM16B_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 8) | NC score | 0.675992 (rank : 8) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQ90 | Gene names | TMEM16B, C12orf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protein 16B. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 9) | NC score | 0.059257 (rank : 40) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.061844 (rank : 34) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 11) | NC score | 0.093540 (rank : 9) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 12) | NC score | 0.049864 (rank : 83) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.057109 (rank : 48) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.082033 (rank : 13) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
EFS_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.072930 (rank : 17) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.047088 (rank : 90) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
ADCY8_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.054407 (rank : 63) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.055792 (rank : 51) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ADCY8_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.053465 (rank : 68) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.047865 (rank : 86) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
EPHB6_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.012029 (rank : 159) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08644, Q8BN76, Q8K0A9 | Gene names | Ephb6, Cekl | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (MEP). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.063751 (rank : 30) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.054553 (rank : 61) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.061465 (rank : 37) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
IER5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.071330 (rank : 18) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.035586 (rank : 105) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.048846 (rank : 85) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NUCB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.050723 (rank : 78) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.018394 (rank : 140) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.063149 (rank : 31) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
AMOL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.038512 (rank : 97) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.047262 (rank : 89) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
AATK_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.014877 (rank : 148) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.057845 (rank : 44) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.025081 (rank : 125) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.025591 (rank : 123) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.057501 (rank : 46) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.028066 (rank : 119) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.036038 (rank : 104) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.019739 (rank : 137) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.033900 (rank : 108) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.054796 (rank : 59) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.054541 (rank : 62) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ZN710_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.001916 (rank : 183) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.047701 (rank : 87) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.031159 (rank : 116) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.024549 (rank : 128) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.049677 (rank : 84) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.021425 (rank : 134) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.012484 (rank : 157) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.043665 (rank : 94) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
LRC56_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.038251 (rank : 98) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
NELF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.033146 (rank : 112) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6X4W1, Q6X4V7, Q6X4V8, Q6X4V9, Q8N2M2, Q96SY1, Q9NPM4, Q9NPP3, Q9NPS3 | Gene names | NELF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.031810 (rank : 115) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
C1QA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.027651 (rank : 120) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02745, Q5T963 | Gene names | C1QA | |||
|
Domain Architecture |
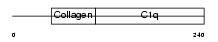
|
|||||
| Description | Complement C1q subcomponent subunit A precursor. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.045653 (rank : 92) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.013887 (rank : 152) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.054940 (rank : 57) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.036918 (rank : 101) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.035421 (rank : 106) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.023775 (rank : 130) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.017496 (rank : 144) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
AMOL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.033446 (rank : 111) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
ATS12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.009016 (rank : 170) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
CC036_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.047281 (rank : 88) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3SXR2, Q3SXR3, Q9H6K8 | Gene names | C3orf36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf36. | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.051445 (rank : 74) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.045517 (rank : 93) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.033840 (rank : 109) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
EPHB6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.009730 (rank : 169) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15197 | Gene names | EPHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (HEP). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.003814 (rank : 180) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.005912 (rank : 175) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.012873 (rank : 153) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.025024 (rank : 126) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.034245 (rank : 107) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.032449 (rank : 113) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.036968 (rank : 100) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ANR47_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.012607 (rank : 154) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
ATG9A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.025584 (rank : 124) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
CO039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.064111 (rank : 29) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
CO039_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.053335 (rank : 70) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.028423 (rank : 118) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
KLF8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.001222 (rank : 185) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95600, Q59GV5, Q5HYQ5, Q5JXP7, Q6MZJ7, Q9UGC4 | Gene names | KLF8, BKLF3, ZNF741 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 8 (Basic krueppel-like factor 3) (Zinc finger protein 741). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.040174 (rank : 96) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.065765 (rank : 26) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
AF10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.023945 (rank : 129) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.033726 (rank : 110) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.014390 (rank : 151) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.017971 (rank : 142) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MARK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.003804 (rank : 181) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.050234 (rank : 80) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PCD12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.002702 (rank : 182) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PELI3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.019659 (rank : 138) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N2H9, Q8N3E1, Q8N9Q6, Q8TAW7, Q8TED5 | Gene names | PELI3 | |||
|
Domain Architecture |
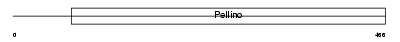
|
|||||
| Description | Protein pellino homolog 3 (Pellino-3). | |||||
|
PELI3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.019639 (rank : 139) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXR6, Q8K329 | Gene names | Peli3 | |||
|
Domain Architecture |
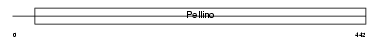
|
|||||
| Description | Protein pellino homolog 3 (Pellino-3). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.004187 (rank : 179) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.000168 (rank : 186) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
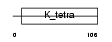
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.025828 (rank : 122) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.019888 (rank : 136) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.026358 (rank : 121) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.024971 (rank : 127) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.022625 (rank : 131) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.037412 (rank : 99) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
EFS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.046245 (rank : 91) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
GAS8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.011721 (rank : 164) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.021570 (rank : 133) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.029137 (rank : 117) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.012441 (rank : 158) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.016319 (rank : 146) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
ASGR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.005067 (rank : 177) | |||
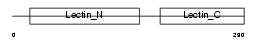
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
BSCL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.041870 (rank : 95) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.055183 (rank : 54) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.036228 (rank : 102) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.005839 (rank : 176) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DNJA3_HUMAN
|
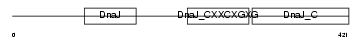
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.006602 (rank : 173) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.010095 (rank : 167) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
JHD3D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.015358 (rank : 147) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
MCPT1_MOUSE
|
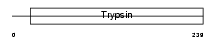
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.001254 (rank : 184) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11034 | Gene names | Mcpt1 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 1 precursor (EC 3.4.21.-) (MMCP-1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.032089 (rank : 114) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.056421 (rank : 49) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.020558 (rank : 135) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PUNC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.006560 (rank : 174) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.012558 (rank : 155) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
SAM14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.017525 (rank : 143) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.010660 (rank : 165) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.004872 (rank : 178) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
DMXL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.006994 (rank : 171) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
FRS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.014825 (rank : 149) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.012540 (rank : 156) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IRX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.011738 (rank : 163) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.018326 (rank : 141) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MIS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.010414 (rank : 166) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.022125 (rank : 132) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.014400 (rank : 150) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.006775 (rank : 172) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.016984 (rank : 145) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.009925 (rank : 168) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.036152 (rank : 103) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.011855 (rank : 161) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TLE4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.011878 (rank : 160) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
TLE4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.011762 (rank : 162) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.054960 (rank : 56) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.069774 (rank : 20) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.078024 (rank : 14) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050198 (rank : 81) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.055883 (rank : 50) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.054915 (rank : 58) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.057709 (rank : 45) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.059159 (rank : 41) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.053197 (rank : 72) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.066734 (rank : 25) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.054650 (rank : 60) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051458 (rank : 73) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.055301 (rank : 53) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.053361 (rank : 69) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.061901 (rank : 33) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.070086 (rank : 19) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.058809 (rank : 42) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.060699 (rank : 39) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.068933 (rank : 21) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.058215 (rank : 43) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.054256 (rank : 64) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.055152 (rank : 55) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.084719 (rank : 12) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.075587 (rank : 16) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.057378 (rank : 47) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.088654 (rank : 10) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.085686 (rank : 11) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.075850 (rank : 15) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.061480 (rank : 36) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.053790 (rank : 66) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.053262 (rank : 71) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SELV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050053 (rank : 82) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.053748 (rank : 67) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050779 (rank : 77) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.050294 (rank : 79) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.064728 (rank : 28) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.061496 (rank : 35) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.060879 (rank : 38) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.067731 (rank : 23) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.055705 (rank : 52) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.053839 (rank : 65) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.068672 (rank : 22) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.067651 (rank : 24) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.050805 (rank : 76) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.065632 (rank : 27) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.062763 (rank : 32) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.051063 (rank : 75) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
TM16H_MOUSE
|
||||||
| NC score | 0.953761 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
TM16E_MOUSE
|
||||||
| NC score | 0.689390 (rank : 3) | θ value | 4.73814e-25 (rank : 3) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q75UR0, Q8CC04 | Gene names | Tmem16e, Gdd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16E (Gnathodiaphyseal dysplasia 1 protein homolog). | |||||
|
TM16F_MOUSE
|
||||||
| NC score | 0.686335 (rank : 4) | θ value | 8.93572e-24 (rank : 4) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P9J9, Q8C242 | Gene names | Tmem16f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16F. | |||||
|
TM16F_HUMAN
|
||||||
| NC score | 0.684819 (rank : 5) | θ value | 4.43474e-23 (rank : 6) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q4KMQ2, Q8N3Q2 | Gene names | TMEM16F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16F. | |||||
|
TM16C_HUMAN
|
||||||
| NC score | 0.683996 (rank : 6) | θ value | 6.40375e-22 (rank : 7) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYT9 | Gene names | TMEM16C, C11orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protein 16C. | |||||
|
TM16E_HUMAN
|
||||||
| NC score | 0.683896 (rank : 7) | θ value | 1.99067e-23 (rank : 5) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q75V66 | Gene names | TMEM16E, GDD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16E (Gnathodiaphyseal dysplasia 1 protein). | |||||
|
TM16B_HUMAN
|
||||||
| NC score | 0.675992 (rank : 8) | θ value | 8.65492e-19 (rank : 8) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQ90 | Gene names | TMEM16B, C12orf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protein 16B. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.093540 (rank : 9) | θ value | 0.00134147 (rank : 11) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.088654 (rank : 10) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.085686 (rank : 11) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.084719 (rank : 12) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.082033 (rank : 13) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.078024 (rank : 14) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.075850 (rank : 15) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.075587 (rank : 16) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.072930 (rank : 17) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
IER5_MOUSE
|
||||||
| NC score | 0.071330 (rank : 18) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.070086 (rank : 19) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.069774 (rank : 20) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.068933 (rank : 21) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.068672 (rank : 22) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.067731 (rank : 23) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.067651 (rank : 24) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.066734 (rank : 25) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.065765 (rank : 26) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.065632 (rank : 27) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.064728 (rank : 28) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.064111 (rank : 29) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.063751 (rank : 30) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.063149 (rank : 31) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.062763 (rank : 32) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.061901 (rank : 33) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.061844 (rank : 34) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.061496 (rank : 35) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.061480 (rank : 36) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.061465 (rank : 37) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.060879 (rank : 38) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.060699 (rank : 39) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.059257 (rank : 40) | θ value | 4.1701e-05 (rank : 9) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.059159 (rank : 41) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.058809 (rank : 42) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.058215 (rank : 43) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.057845 (rank : 44) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.057709 (rank : 45) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.057501 (rank : 46) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.057378 (rank : 47) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.057109 (rank : 48) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.056421 (rank : 49) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.055883 (rank : 50) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055792 (rank : 51) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.055705 (rank : 52) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.055301 (rank : 53) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.055183 (rank : 54) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.055152 (rank : 55) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.054960 (rank : 56) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.054940 (rank : 57) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.054915 (rank : 58) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.054796 (rank : 59) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.054650 (rank : 60) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.054553 (rank : 61) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.054541 (rank : 62) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ADCY8_HUMAN
|
||||||
| NC score | 0.054407 (rank : 63) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.054256 (rank : 64) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.053839 (rank : 65) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.053790 (rank : 66) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.053748 (rank : 67) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ADCY8_MOUSE
|
||||||
| NC score | 0.053465 (rank : 68) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.053361 (rank : 69) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.053335 (rank : 70) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.053262 (rank : 71) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.053197 (rank : 72) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.051458 (rank : 73) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.051445 (rank : 74) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.051063 (rank : 75) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.050805 (rank : 76) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.050779 (rank : 77) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
NUCB1_MOUSE
|
||||||
| NC score | 0.050723 (rank : 78) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.050294 (rank : 79) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.050234 (rank : 80) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.050198 (rank : 81) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.050053 (rank : 82) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.049864 (rank : 83) | θ value | 0.00175202 (rank : 12) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.049677 (rank : 84) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.048846 (rank : 85) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.047865 (rank : 86) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.047701 (rank : 87) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CC036_HUMAN
|
||||||
| NC score | 0.047281 (rank : 88) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3SXR2, Q3SXR3, Q9H6K8 | Gene names | C3orf36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf36. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.047262 (rank : 89) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.047088 (rank : 90) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.046245 (rank : 91) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.045653 (rank : 92) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.045517 (rank : 93) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.043665 (rank : 94) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
BSCL2_MOUSE
|
||||||
| NC score | 0.041870 (rank : 95) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.040174 (rank : 96) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
AMOL1_HUMAN
|
||||||
| NC score | 0.038512 (rank : 97) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY63, Q63HK7, Q8NDN0, Q8WXD1, Q96CM5 | Gene names | AMOTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
LRC56_HUMAN
|
||||||
| NC score | 0.038251 (rank : 98) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.037412 (rank : 99) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.036968 (rank : 100) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.036918 (rank : 101) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.036228 (rank : 102) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.036152 (rank : 103) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.036038 (rank : 104) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.035586 (rank : 105) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.035421 (rank : 106) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.034245 (rank : 107) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.033900 (rank : 108) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.033840 (rank : 109) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.033726 (rank : 110) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
AMOL1_MOUSE
|
||||||
| NC score | 0.033446 (rank : 111) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
NELF_HUMAN
|
||||||
| NC score | 0.033146 (rank : 112) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6X4W1, Q6X4V7, Q6X4V8, Q6X4V9, Q8N2M2, Q96SY1, Q9NPM4, Q9NPP3, Q9NPS3 | Gene names | NELF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor). | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.032449 (rank : 113) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.032089 (rank : 114) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.031810 (rank : 115) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.031159 (rank : 116) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.029137 (rank : 117) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.028423 (rank : 118) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.028066 (rank : 119) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
C1QA_HUMAN
|
||||||
| NC score | 0.027651 (rank : 120) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02745, Q5T963 | Gene names | C1QA | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1q subcomponent subunit A precursor. | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.026358 (rank : 121) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.025828 (rank : 122) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.025591 (rank : 123) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
ATG9A_HUMAN
|
||||||
| NC score | 0.025584 (rank : 124) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.025081 (rank : 125) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.025024 (rank : 126) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.024971 (rank : 127) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.024549 (rank : 128) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.023945 (rank : 129) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.023775 (rank : 130) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.022625 (rank : 131) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.022125 (rank : 132) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.021570 (rank : 133) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.021425 (rank : 134) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.020558 (rank : 135) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.019888 (rank : 136) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.019739 (rank : 137) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PELI3_HUMAN
|
||||||
| NC score | 0.019659 (rank : 138) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N2H9, Q8N3E1, Q8N9Q6, Q8TAW7, Q8TED5 | Gene names | PELI3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 3 (Pellino-3). | |||||
|
PELI3_MOUSE
|
||||||
| NC score | 0.019639 (rank : 139) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXR6, Q8K329 | Gene names | Peli3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 3 (Pellino-3). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.018394 (rank : 140) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.018326 (rank : 141) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.017971 (rank : 142) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
SAM14_HUMAN
|
||||||
| NC score | 0.017525 (rank : 143) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.017496 (rank : 144) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.016984 (rank : 145) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.016319 (rank : 146) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
JHD3D_HUMAN
|
||||||
| NC score | 0.015358 (rank : 147) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.014877 (rank : 148) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
FRS3_MOUSE
|
||||||
| NC score | 0.014825 (rank : 149) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.014400 (rank : 150) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.014390 (rank : 151) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.013887 (rank : 152) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.012873 (rank : 153) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ANR47_MOUSE
|
||||||
| NC score | 0.012607 (rank : 154) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.012558 (rank : 155) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.012540 (rank : 156) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.012484 (rank : 157) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.012441 (rank : 158) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
EPHB6_MOUSE
|
||||||
| NC score | 0.012029 (rank : 159) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08644, Q8BN76, Q8K0A9 | Gene names | Ephb6, Cekl | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (MEP). | |||||
|
TLE4_HUMAN
|
||||||
| NC score | 0.011878 (rank : 160) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.011855 (rank : 161) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TLE4_MOUSE
|
||||||
| NC score | 0.011762 (rank : 162) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
IRX2_HUMAN
|
||||||
| NC score | 0.011738 (rank : 163) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
GAS8_MOUSE
|
||||||
| NC score | 0.011721 (rank : 164) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.010660 (rank : 165) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
MIS_HUMAN
|
||||||
| NC score | 0.010414 (rank : 166) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.010095 (rank : 167) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.009925 (rank : 168) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
EPHB6_HUMAN
|
||||||
| NC score | 0.009730 (rank : 169) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15197 | Gene names | EPHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (HEP). | |||||
|
ATS12_HUMAN
|
||||||
| NC score | 0.009016 (rank : 170) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
DMXL2_HUMAN
|
||||||
| NC score | 0.006994 (rank : 171) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.006775 (rank : 172) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.006602 (rank : 173) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
PUNC_MOUSE
|
||||||
| NC score | 0.006560 (rank : 174) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.005912 (rank : 175) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.005839 (rank : 176) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
ASGR2_MOUSE
|
||||||
| NC score | 0.005067 (rank : 177) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.004872 (rank : 178) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.004187 (rank : 179) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.003814 (rank : 180) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MARK2_MOUSE
|
||||||
| NC score | 0.003804 (rank : 181) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
PCD12_HUMAN
|
||||||
| NC score | 0.002702 (rank : 182) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
ZN710_HUMAN
|
||||||
| NC score | 0.001916 (rank : 183) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
MCPT1_MOUSE
|
||||||
| NC score | 0.001254 (rank : 184) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11034 | Gene names | Mcpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 1 precursor (EC 3.4.21.-) (MMCP-1). | |||||
|
KLF8_HUMAN
|
||||||
| NC score | 0.001222 (rank : 185) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95600, Q59GV5, Q5HYQ5, Q5JXP7, Q6MZJ7, Q9UGC4 | Gene names | KLF8, BKLF3, ZNF741 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 8 (Basic krueppel-like factor 3) (Zinc finger protein 741). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | 0.000168 (rank : 186) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||