Please be patient as the page loads
|
ZCPW1_HUMAN
|
||||||
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 2.04937e-153 (rank : 2) | NC score | 0.860182 (rank : 2) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
MORC4_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 3) | NC score | 0.314987 (rank : 4) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MORC4_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 4) | NC score | 0.316566 (rank : 3) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 5) | NC score | 0.255176 (rank : 5) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 6) | NC score | 0.252764 (rank : 6) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MORC3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 7) | NC score | 0.230035 (rank : 7) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14149, Q9UEZ2 | Gene names | MORC3, KIAA0136, ZCWCC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 3 (Zinc finger CW-type coiled- coil domain protein 3). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.139867 (rank : 20) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.069254 (rank : 38) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.156158 (rank : 15) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.082819 (rank : 33) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.067577 (rank : 45) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.054649 (rank : 62) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HDGF_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.203271 (rank : 9) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.197328 (rank : 10) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.055969 (rank : 59) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.051601 (rank : 67) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.062660 (rank : 53) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.147855 (rank : 16) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
PWWP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.210265 (rank : 8) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.146355 (rank : 17) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.068785 (rank : 43) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.040109 (rank : 86) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
DNM3B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.098011 (rank : 26) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.022443 (rank : 116) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.056521 (rank : 58) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.084503 (rank : 32) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PSIP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.171520 (rank : 14) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.179054 (rank : 11) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.069203 (rank : 39) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
GCP60_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.049279 (rank : 76) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
MMTA2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.068485 (rank : 44) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
SASH1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.042385 (rank : 84) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.113924 (rank : 23) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.055386 (rank : 61) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
MOR2A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.142589 (rank : 19) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.048243 (rank : 77) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.056638 (rank : 57) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.064383 (rank : 50) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
HDGR3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.178178 (rank : 12) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
HDGR3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.177700 (rank : 13) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
MORC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.146291 (rank : 18) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.092105 (rank : 27) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.098790 (rank : 25) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.028517 (rank : 105) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.065432 (rank : 47) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.051440 (rank : 69) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.029390 (rank : 102) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.029519 (rank : 101) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.055727 (rank : 60) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.035922 (rank : 94) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
AOF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.068858 (rank : 42) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
APOA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.008092 (rank : 134) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.043724 (rank : 83) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.037606 (rank : 88) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.088410 (rank : 29) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
DNM3A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.085168 (rank : 31) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.005615 (rank : 137) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
SOCS5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.025854 (rank : 111) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54928 | Gene names | Socs5, Cish5 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.066054 (rank : 46) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.062302 (rank : 54) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.036851 (rank : 92) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
TM16H_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.032449 (rank : 96) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.020374 (rank : 120) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
DNM3B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.089461 (rank : 28) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.021151 (rank : 119) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
MYO1F_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.007778 (rank : 135) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00160, Q8WWN7 | Gene names | MYO1F | |||
|
Domain Architecture |

|
|||||
| Description | Myosin If (Myosin-IE). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.028868 (rank : 104) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
SRPK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.023076 (rank : 115) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
SRPK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.019733 (rank : 122) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
STMN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.050468 (rank : 72) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |
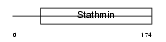
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.050468 (rank : 73) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |
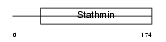
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.032903 (rank : 95) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.046639 (rank : 79) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.029138 (rank : 103) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.045001 (rank : 81) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.031168 (rank : 99) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
LIPH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.014925 (rank : 129) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27656 | Gene names | Lipc, Hpl | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic triacylglycerol lipase precursor (EC 3.1.1.3) (Hepatic lipase) (HL). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.070434 (rank : 37) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAOK3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.010612 (rank : 132) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
WWC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.027064 (rank : 108) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.040518 (rank : 85) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.015688 (rank : 128) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.027711 (rank : 106) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.037210 (rank : 89) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.013251 (rank : 130) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.044131 (rank : 82) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.027192 (rank : 107) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CT116_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.026642 (rank : 109) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80WW9, Q9CT52 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 homolog precursor. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.030769 (rank : 100) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
KCTD8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.011398 (rank : 131) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KPCT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | -0.000276 (rank : 139) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04759, Q3MJF1, Q64FY5, Q9H508, Q9H549 | Gene names | PRKCQ, PRKCT | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.025535 (rank : 113) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.086271 (rank : 30) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.036735 (rank : 93) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.039064 (rank : 87) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.021468 (rank : 117) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
PRPF3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.017789 (rank : 127) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.005334 (rank : 138) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.009881 (rank : 133) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.046115 (rank : 80) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.031665 (rank : 98) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CI005_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.020260 (rank : 121) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H330, Q5JTQ5, Q5SS43, Q6ZME3, Q8NDJ5, Q96CG6 | Gene names | C9orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf5 (Protein CG-2). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.021444 (rank : 118) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.036947 (rank : 90) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.026435 (rank : 110) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.018403 (rank : 126) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
NDF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.006990 (rank : 136) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
OXR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.025561 (rank : 112) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
P80C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.032085 (rank : 97) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.025509 (rank : 114) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.019266 (rank : 123) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SNX4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.018978 (rank : 125) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95219 | Gene names | SNX4 | |||
|
Domain Architecture |
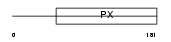
|
|||||
| Description | Sorting nexin-4. | |||||
|
SNX4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.019211 (rank : 124) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YJ2 | Gene names | Snx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-4. | |||||
|
STMN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.036905 (rank : 91) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |
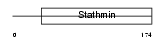
|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.046821 (rank : 78) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.058097 (rank : 56) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.064027 (rank : 51) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.052165 (rank : 65) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052027 (rank : 66) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
ENL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.050243 (rank : 75) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
K1143_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.050786 (rank : 70) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96AT1, Q96HJ8, Q9ULS7 | Gene names | KIAA1143 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143. | |||||
|
MOR2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.116791 (rank : 22) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
MORC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.111781 (rank : 24) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86VD1, Q7L8E2, Q9NSG7, Q9Y6D4 | Gene names | MORC1, MORC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1. | |||||
|
MORC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.120086 (rank : 21) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WVL5 | Gene names | Morc1, Morc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1 (Protein microrchidia). | |||||
|
MSH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.065203 (rank : 48) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.063422 (rank : 52) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.068910 (rank : 41) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
MSH3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.069156 (rank : 40) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.058205 (rank : 55) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
MSH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.050372 (rank : 74) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
MSH5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.054223 (rank : 63) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.053377 (rank : 64) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.074296 (rank : 35) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050471 (rank : 71) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.077408 (rank : 34) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.051582 (rank : 68) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.071609 (rank : 36) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.064765 (rank : 49) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.860182 (rank : 2) | θ value | 2.04937e-153 (rank : 2) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
MORC4_MOUSE
|
||||||
| NC score | 0.316566 (rank : 3) | θ value | 6.87365e-08 (rank : 4) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MORC4_HUMAN
|
||||||
| NC score | 0.314987 (rank : 4) | θ value | 1.06045e-08 (rank : 3) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.255176 (rank : 5) | θ value | 5.81887e-07 (rank : 5) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_HUMAN
|
||||||
| NC score | 0.252764 (rank : 6) | θ value | 7.59969e-07 (rank : 6) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MORC3_HUMAN
|
||||||
| NC score | 0.230035 (rank : 7) | θ value | 1.87187e-05 (rank : 7) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14149, Q9UEZ2 | Gene names | MORC3, KIAA0136, ZCWCC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 3 (Zinc finger CW-type coiled- coil domain protein 3). | |||||
|
PWWP2_HUMAN
|
||||||
| NC score | 0.210265 (rank : 8) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
HDGF_HUMAN
|
||||||
| NC score | 0.203271 (rank : 9) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.197328 (rank : 10) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.179054 (rank : 11) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
HDGR3_HUMAN
|
||||||
| NC score | 0.178178 (rank : 12) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
HDGR3_MOUSE
|
||||||
| NC score | 0.177700 (rank : 13) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
PSIP1_HUMAN
|
||||||
| NC score | 0.171520 (rank : 14) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.156158 (rank : 15) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.147855 (rank : 16) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.146355 (rank : 17) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
MORC2_HUMAN
|
||||||
| NC score | 0.146291 (rank : 18) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
MOR2A_MOUSE
|
||||||
| NC score | 0.142589 (rank : 19) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.139867 (rank : 20) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
MORC1_MOUSE
|
||||||
| NC score | 0.120086 (rank : 21) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WVL5 | Gene names | Morc1, Morc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1 (Protein microrchidia). | |||||
|
MOR2B_MOUSE
|
||||||
| NC score | 0.116791 (rank : 22) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.113924 (rank : 23) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MORC1_HUMAN
|
||||||
| NC score | 0.111781 (rank : 24) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86VD1, Q7L8E2, Q9NSG7, Q9Y6D4 | Gene names | MORC1, MORC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.098790 (rank : 25) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
DNM3B_MOUSE
|
||||||
| NC score | 0.098011 (rank : 26) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.092105 (rank : 27) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
DNM3B_HUMAN
|
||||||
| NC score | 0.089461 (rank : 28) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.088410 (rank : 29) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.086271 (rank : 30) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
DNM3A_MOUSE
|
||||||
| NC score | 0.085168 (rank : 31) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.084503 (rank : 32) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.082819 (rank : 33) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.077408 (rank : 34) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.074296 (rank : 35) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.071609 (rank : 36) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.070434 (rank : 37) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.069254 (rank : 38) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.069203 (rank : 39) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
MSH3_MOUSE
|
||||||
| NC score | 0.069156 (rank : 40) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH3_HUMAN
|
||||||
| NC score | 0.068910 (rank : 41) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
AOF1_HUMAN
|
||||||
| NC score | 0.068858 (rank : 42) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.068785 (rank : 43) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MMTA2_MOUSE
|
||||||
| NC score | 0.068485 (rank : 44) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.067577 (rank : 45) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.066054 (rank : 46) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.065432 (rank : 47) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MSH2_HUMAN
|
||||||
| NC score | 0.065203 (rank : 48) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.064765 (rank : 49) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.064383 (rank : 50) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.064027 (rank : 51) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
MSH2_MOUSE
|
||||||
| NC score | 0.063422 (rank : 52) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.062660 (rank : 53) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.062302 (rank : 54) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
MSH4_HUMAN
|
||||||
| NC score | 0.058205 (rank : 55) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.058097 (rank : 56) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.056638 (rank : 57) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.056521 (rank : 58) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.055969 (rank : 59) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.055727 (rank : 60) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.055386 (rank : 61) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.054649 (rank : 62) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MSH5_HUMAN
|
||||||
| NC score | 0.054223 (rank : 63) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_MOUSE
|
||||||
| NC score | 0.053377 (rank : 64) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.052165 (rank : 65) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.052027 (rank : 66) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.051601 (rank : 67) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.051582 (rank : 68) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.051440 (rank : 69) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
K1143_HUMAN
|
||||||
| NC score | 0.050786 (rank : 70) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q96AT1, Q96HJ8, Q9ULS7 | Gene names | KIAA1143 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143. | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.050471 (rank : 71) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
STMN2_HUMAN
|
||||||
| NC score | 0.050468 (rank : 72) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| NC score | 0.050468 (rank : 73) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
MSH4_MOUSE
|
||||||
| NC score | 0.050372 (rank : 74) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.050243 (rank : 75) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
GCP60_MOUSE
|
||||||
| NC score | 0.049279 (rank : 76) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.048243 (rank : 77) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.046821 (rank : 78) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.046639 (rank : 79) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.046115 (rank : 80) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.045001 (rank : 81) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.044131 (rank : 82) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.043724 (rank : 83) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
SASH1_MOUSE
|
||||||
| NC score | 0.042385 (rank : 84) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.040518 (rank : 85) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.040109 (rank : 86) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.039064 (rank : 87) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.037606 (rank : 88) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.037210 (rank : 89) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.036947 (rank : 90) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
STMN3_HUMAN
|
||||||
| NC score | 0.036905 (rank : 91) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.036851 (rank : 92) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.036735 (rank : 93) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.035922 (rank : 94) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.032903 (rank : 95) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.032449 (rank : 96) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.032085 (rank : 97) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.031665 (rank : 98) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.031168 (rank : 99) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.030769 (rank : 100) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.029519 (rank : 101) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.029390 (rank : 102) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.029138 (rank : 103) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.028868 (rank : 104) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.028517 (rank : 105) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.027711 (rank : 106) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.027192 (rank : 107) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
WWC3_HUMAN
|
||||||
| NC score | 0.027064 (rank : 108) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
CT116_MOUSE
|
||||||
| NC score | 0.026642 (rank : 109) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80WW9, Q9CT52 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 homolog precursor. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.026435 (rank : 110) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
SOCS5_MOUSE
|
||||||
| NC score | 0.025854 (rank : 111) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54928 | Gene names | Socs5, Cish5 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5). | |||||
|
OXR1_HUMAN
|
||||||
| NC score | 0.025561 (rank : 112) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.025535 (rank : 113) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.025509 (rank : 114) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SRPK1_HUMAN
|
||||||
| NC score | 0.023076 (rank : 115) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.022443 (rank : 116) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.021468 (rank : 117) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.021444 (rank : 118) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.021151 (rank : 119) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.020374 (rank : 120) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CI005_HUMAN
|
||||||
| NC score | 0.020260 (rank : 121) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H330, Q5JTQ5, Q5SS43, Q6ZME3, Q8NDJ5, Q96CG6 | Gene names | C9orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf5 (Protein CG-2). | |||||
|
SRPK1_MOUSE
|
||||||
| NC score | 0.019733 (rank : 122) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.019266 (rank : 123) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SNX4_MOUSE
|
||||||
| NC score | 0.019211 (rank : 124) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YJ2 | Gene names | Snx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-4. | |||||
|
SNX4_HUMAN
|
||||||
| NC score | 0.018978 (rank : 125) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95219 | Gene names | SNX4 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-4. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.018403 (rank : 126) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
PRPF3_MOUSE
|
||||||
| NC score | 0.017789 (rank : 127) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.015688 (rank : 128) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
LIPH_MOUSE
|
||||||
| NC score | 0.014925 (rank : 129) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27656 | Gene names | Lipc, Hpl | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic triacylglycerol lipase precursor (EC 3.1.1.3) (Hepatic lipase) (HL). | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.013251 (rank : 130) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
KCTD8_MOUSE
|
||||||
| NC score | 0.011398 (rank : 131) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
TAOK3_MOUSE
|
||||||
| NC score | 0.010612 (rank : 132) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.009881 (rank : 133) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
APOA_HUMAN
|
||||||
| NC score | 0.008092 (rank : 134) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
MYO1F_HUMAN
|
||||||
| NC score | 0.007778 (rank : 135) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00160, Q8WWN7 | Gene names | MYO1F | |||
|
Domain Architecture |

|
|||||
| Description | Myosin If (Myosin-IE). | |||||
|
NDF2_MOUSE
|
||||||
| NC score | 0.006990 (rank : 136) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.005615 (rank : 137) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.005334 (rank : 138) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
KPCT_HUMAN
|
||||||
| NC score | -0.000276 (rank : 139) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04759, Q3MJF1, Q64FY5, Q9H508, Q9H549 | Gene names | PRKCQ, PRKCT | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||