Please be patient as the page loads
|
MSH6_HUMAN
|
||||||
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MSH6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991263 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH3_MOUSE
|
||||||
| θ value | 1.65806e-78 (rank : 3) | NC score | 0.855877 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH3_HUMAN
|
||||||
| θ value | 2.1655e-78 (rank : 4) | NC score | 0.853283 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
MSH2_HUMAN
|
||||||
| θ value | 3.0428e-56 (rank : 5) | NC score | 0.822710 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH2_MOUSE
|
||||||
| θ value | 3.36421e-55 (rank : 6) | NC score | 0.821127 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH5_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 7) | NC score | 0.754730 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 8) | NC score | 0.753003 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH4_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 9) | NC score | 0.755530 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
MSH4_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 10) | NC score | 0.751674 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 11) | NC score | 0.252764 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 12) | NC score | 0.119524 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 13) | NC score | 0.227892 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 14) | NC score | 0.192523 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
PSIP1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 15) | NC score | 0.182423 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
PWWP2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.195746 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 17) | NC score | 0.042336 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
DNM3B_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 18) | NC score | 0.112596 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
DNM3B_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.110759 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
HDGF_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.177498 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.173942 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
HDGR3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.169029 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.038593 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.045583 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
HDGR3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.167971 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.044096 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.039255 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.094313 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.029255 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.032513 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.024630 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.025356 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.038752 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.030330 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DNM3A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.094091 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
HORN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.021639 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.081592 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.082063 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.030803 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.070009 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.017392 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.049870 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.037751 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
SALL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.002808 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.026562 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
SYMM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.030738 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.032145 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.018867 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.019720 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.005266 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SYMM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.029349 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
CHIC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.035177 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKJ5 | Gene names | CHIC2, BTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 2 protein (BrX-like translocated in leukemia). | |||||
|
CHIC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.035216 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D9G3, Q3T9U2, Q80ZT7 | Gene names | Chic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 2 protein. | |||||
|
GP101_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.001771 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80T62, Q3UUI9 | Gene names | Gpr101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 101. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.037857 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.009390 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.014410 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.016941 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
ZBT24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | -0.000996 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
BEGIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.010907 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
CIR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.024643 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
DCX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.010211 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
KIFC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.003201 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BW19, O60887, Q14834, Q9UQP7 | Gene names | KIFC1, HSET, KNSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC1 (Kinesin-like protein 2) (Kinesin-related protein HSET). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.009288 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.016352 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.022850 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TYRO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.015079 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11344 | Gene names | Tyr | |||
|
Domain Architecture |
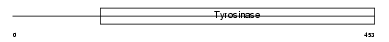
|
|||||
| Description | Tyrosinase precursor (EC 1.14.18.1) (Monophenol monooxygenase) (Albino locus protein). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.015323 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.025857 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.005852 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.004467 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.015186 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RMP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.024725 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94763 | Gene names | RMP, C19orf2 | |||
|
Domain Architecture |
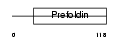
|
|||||
| Description | RNA polymerase II subunit 5-mediating protein (RPB5-mediating protein). | |||||
|
ANXA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.003707 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08133, Q6ZT79 | Gene names | ANXA6, ANX6 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A6 (Annexin VI) (Lipocortin VI) (P68) (P70) (Protein III) (Chromobindin-20) (67 kDa calelectrin) (Calphobindin-II) (CPB-II). | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.005337 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
K0056_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.010051 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.021668 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.024450 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
SYSM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.009195 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP81, Q9BVP3 | Gene names | SARS2, SARSM | |||
|
Domain Architecture |
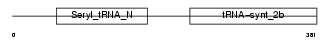
|
|||||
| Description | Seryl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.11) (Serine-- tRNA ligase) (SerRSmt). | |||||
|
TALDO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.011279 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37837, O00751 | Gene names | TALDO1, TAL, TALDO, TALDOR | |||
|
Domain Architecture |

|
|||||
| Description | Transaldolase (EC 2.2.1.2). | |||||
|
ZN398_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | -0.001943 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TD17, Q8TD18, Q9P2K7, Q9UDV8 | Gene names | ZNF398, KIAA1339, ZER6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 398 (Zinc finger DNA-binding protein p52/p71). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052279 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
MSH6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.991263 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH3_MOUSE
|
||||||
| NC score | 0.855877 (rank : 3) | θ value | 1.65806e-78 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH3_HUMAN
|
||||||
| NC score | 0.853283 (rank : 4) | θ value | 2.1655e-78 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
MSH2_HUMAN
|
||||||
| NC score | 0.822710 (rank : 5) | θ value | 3.0428e-56 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH2_MOUSE
|
||||||
| NC score | 0.821127 (rank : 6) | θ value | 3.36421e-55 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH4_HUMAN
|
||||||
| NC score | 0.755530 (rank : 7) | θ value | 8.3442e-30 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
MSH5_HUMAN
|
||||||
| NC score | 0.754730 (rank : 8) | θ value | 1.16434e-31 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_MOUSE
|
||||||
| NC score | 0.753003 (rank : 9) | θ value | 2.59387e-31 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH4_MOUSE
|
||||||
| NC score | 0.751674 (rank : 10) | θ value | 1.4233e-29 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.252764 (rank : 11) | θ value | 7.59969e-07 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.227892 (rank : 12) | θ value | 9.92553e-07 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
PWWP2_HUMAN
|
||||||
| NC score | 0.195746 (rank : 13) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.192523 (rank : 14) | θ value | 1.43324e-05 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
PSIP1_HUMAN
|
||||||
| NC score | 0.182423 (rank : 15) | θ value | 0.000461057 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
HDGF_HUMAN
|
||||||
| NC score | 0.177498 (rank : 16) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.173942 (rank : 17) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
HDGR3_HUMAN
|
||||||
| NC score | 0.169029 (rank : 18) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
HDGR3_MOUSE
|
||||||
| NC score | 0.167971 (rank : 19) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.119524 (rank : 20) | θ value | 9.92553e-07 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
DNM3B_HUMAN
|
||||||
| NC score | 0.112596 (rank : 21) | θ value | 0.0148317 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
DNM3B_MOUSE
|
||||||
| NC score | 0.110759 (rank : 22) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.094313 (rank : 23) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
DNM3A_MOUSE
|
||||||
| NC score | 0.094091 (rank : 24) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.082063 (rank : 25) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.081592 (rank : 26) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.070009 (rank : 27) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.052279 (rank : 28) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.049870 (rank : 29) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.045583 (rank : 30) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.044096 (rank : 31) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.042336 (rank : 32) | θ value | 0.0113563 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.039255 (rank : 33) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.038752 (rank : 34) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.038593 (rank : 35) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.037857 (rank : 36) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.037751 (rank : 37) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CHIC2_MOUSE
|
||||||
| NC score | 0.035216 (rank : 38) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D9G3, Q3T9U2, Q80ZT7 | Gene names | Chic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 2 protein. | |||||
|
CHIC2_HUMAN
|
||||||
| NC score | 0.035177 (rank : 39) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKJ5 | Gene names | CHIC2, BTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 2 protein (BrX-like translocated in leukemia). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.032513 (rank : 40) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.032145 (rank : 41) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.030803 (rank : 42) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
SYMM_HUMAN
|
||||||
| NC score | 0.030738 (rank : 43) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.030330 (rank : 44) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
SYMM_MOUSE
|
||||||
| NC score | 0.029349 (rank : 45) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.029255 (rank : 46) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.026562 (rank : 47) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.025857 (rank : 48) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.025356 (rank : 49) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
RMP_HUMAN
|
||||||
| NC score | 0.024725 (rank : 50) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94763 | Gene names | RMP, C19orf2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit 5-mediating protein (RPB5-mediating protein). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.024643 (rank : 51) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.024630 (rank : 52) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.024450 (rank : 53) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.022850 (rank : 54) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.021668 (rank : 55) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.021639 (rank : 56) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.019720 (rank : 57) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.018867 (rank : 58) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.017392 (rank : 59) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.016941 (rank : 60) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.016352 (rank : 61) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.015323 (rank : 62) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.015186 (rank : 63) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TYRO_MOUSE
|
||||||
| NC score | 0.015079 (rank : 64) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11344 | Gene names | Tyr | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosinase precursor (EC 1.14.18.1) (Monophenol monooxygenase) (Albino locus protein). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.014410 (rank : 65) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
TALDO_HUMAN
|
||||||
| NC score | 0.011279 (rank : 66) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37837, O00751 | Gene names | TALDO1, TAL, TALDO, TALDOR | |||
|
Domain Architecture |

|
|||||
| Description | Transaldolase (EC 2.2.1.2). | |||||
|
BEGIN_HUMAN
|
||||||
| NC score | 0.010907 (rank : 67) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.010211 (rank : 68) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
K0056_MOUSE
|
||||||
| NC score | 0.010051 (rank : 69) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.009390 (rank : 70) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.009288 (rank : 71) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SYSM_HUMAN
|
||||||
| NC score | 0.009195 (rank : 72) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP81, Q9BVP3 | Gene names | SARS2, SARSM | |||
|
Domain Architecture |

|
|||||
| Description | Seryl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.11) (Serine-- tRNA ligase) (SerRSmt). | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.005852 (rank : 73) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | 0.005337 (rank : 74) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.005266 (rank : 75) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.004467 (rank : 76) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
ANXA6_HUMAN
|
||||||
| NC score | 0.003707 (rank : 77) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08133, Q6ZT79 | Gene names | ANXA6, ANX6 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A6 (Annexin VI) (Lipocortin VI) (P68) (P70) (Protein III) (Chromobindin-20) (67 kDa calelectrin) (Calphobindin-II) (CPB-II). | |||||
|
KIFC1_HUMAN
|
||||||
| NC score | 0.003201 (rank : 78) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BW19, O60887, Q14834, Q9UQP7 | Gene names | KIFC1, HSET, KNSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC1 (Kinesin-like protein 2) (Kinesin-related protein HSET). | |||||
|
SALL1_HUMAN
|
||||||
| NC score | 0.002808 (rank : 79) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
GP101_MOUSE
|
||||||
| NC score | 0.001771 (rank : 80) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80T62, Q3UUI9 | Gene names | Gpr101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 101. | |||||
|
ZBT24_HUMAN
|
||||||
| NC score | -0.000996 (rank : 81) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
ZN398_HUMAN
|
||||||
| NC score | -0.001943 (rank : 82) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TD17, Q8TD18, Q9P2K7, Q9UDV8 | Gene names | ZNF398, KIAA1339, ZER6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 398 (Zinc finger DNA-binding protein p52/p71). | |||||